In RNA interference, small interfering RNAs recognize messenger RNA homologs in cells and induce their degradation.
When developing siRNA-based drugs for therapeutic use, problems encountered are their low efficiency of delivery to targeted cells and degradation by cellular nucleases. A promising approach to improve cellular delivery of siRNA is bioconjugation. Like other oligonucleotides, siRNA conjugation to lipophilic molecules, antibodies, aptamers, ligands, peptides, or polymers enhances cellular delivery and selective targeting.

Figure 1: Mechanism of systemic and transitive RNAi. Dicer cleavage of dsRNA into 21-23 nt siRNA initiates RNAi. RISC unwinds siRNA duplexes. siRNA binds to mRNA. Either the targeted mRNA is destructed or amplified via RdRP. Amplified dsRNA can transitively silence a 5’-located mRNA in a second cell {Engelke, David R.: RNA interference (RNAi). Nuts and bolts of RNAi technology. DNA Press 2003}.
Modifications of the ribose unit in DNA and RNA oligonucleotides influence the biological properties and base-pairing ability of nucleic acids. In natural RNA, during RNA maturation, various enzymes can introduce chemical modifications into ribonucleotide residues. Chemical alteration can occur in the base, at the 2’-hydroxyl of the ribose, or both. Additional modifications can be introduced chemically in non-natural synthetic RNAs.
The modification of the ribofuranose in nucleic acids allows the manipulation of nucleic acid activities. The local conformation and chemical reactivity of the sugar is changes according to the modification type. Conformational restriction of the ribose units allows the increase in DNA and RNA affinity and enhances biological stability and antisense properties. Alterations in the sugar moiety conformation modify the local and potentially the global structure of nucleic acids. The properties of modified sugar units in oligonucleotides influence the recognition, binding of ligands, and enzymatic activity of proteins.
For example, 2’-modification often improves nuclease resistance, whereas the introduction of bridged nucleic acids (BNAs) significantly increases thermal stability.
Modern automated oligonucleotide synthesis methods enable technologies for the development of plasmid-based gene therapies, antisense, short interfering RNA (siRNA), short hairpin RNA (shRNA), aptamers, and other nucleic acid-based therapeutics. Since natural DNA or RNA oligonucleotides degrade easily, have low binding affinities and poor cellular uptake, chemical modifications overcome many of these limitations and allow the introduction of unique properties as well.
Many chemical modifications have been made to bases, the ribose sugar and the sugar-phosphodiester backbone with the goal to improve their properties for a whole range of applications.
|
A
|
.jpg)
B
|
C
|
Figure 1: siRNA modification sites. A. Labeling nomenclature for ribose sugar unit and phosphate group. Sugar carbons are labeled as 1’-5’ and backbone torsion angles are described as follows: α=O3′(i−1)−P−O5′−C5′; β=P−O5′−C5′−C4′; γ=O5′−C5′−C4′−C3′; δ=C5′−C4′−C3′−O3′; ε=C4′−C3′−O3′−P(i+1); ζ=C3′−O3′−P(i+1)−O5′(i+1) B. Possible sites of introduction of chemical modifications in the sugar moiety and phosphate linkage are marked in red. C. Possible sites for chemical modifications in nucleic acid bases.
Table 1: RNAi modifications of Sugar Units
|
Modification
|
Structure
|
ΔTm duplex per modification
|
Impact on the efficiency of RNAi
|
Others
|
|
2 ′ -O-methyl
(2′O-Me)
|
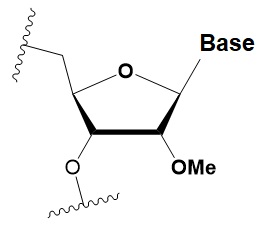
|
+0.5–1.5◦C
|
Two or more consecutive 2′O-Me inhibit RNAi.
Some siRNAs containing 75–82% 2 ′O-Me are biologically active.
|
Stabilizes 3′ endo ribose conformation. ≥5–30% of 2′O-Me increase nuclease resistance in vitro and in vivo.
2 ′O-Me analogs of A, G and U reduce the immune response.
|
|
2 ′ -fluoro
(2′F)
|
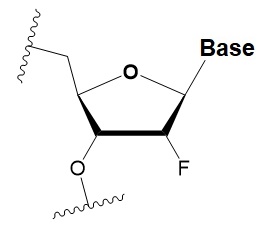
|
+1.5–4◦C
|
2′F analogs in all siRNA positions only slightly reduces the activity of RNAi.
|
Stabilizes 3′ endo ribose conformation.
≥50% 2′F increase nuclease resistance in vitro and in vivo.
2 ′F analogs of adenine (≥7%) reduce the immune response in vitro.
>50% of the 2′F in siRNA may cause toxicity.
|
|
2 ′F-arabinonucleic acid
(2′FANA)
|

|
+1.2◦C
|
100% 2′FANA in the sense chain reduce the efficiency of RNAi.
≥30% 2 ′FANA in the antisense chain inhibit RNAi.
|
Stabilizes 2′ endo ribose conformation.
≥50% 2′FANA increases nuclease resistance in vitro more effectively than 2′F.
Protect siRNA from the action of exoribonucleases.
|
|
2 ′ -O-methoxyethyl
(2′O-MOE)
|
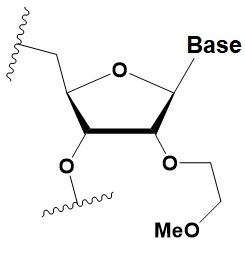
|
+0.9–1.7◦C
|
2′ -MOE at the flanks of the sense strand and the central part (6–11) of the antisense strand are tolerable for RNAi. Replacement of 9th or 10th nucleotides from the 5′ end to 2 ′O-MOE analogs of nucleotide increases the probability of entry in RISC.
|
Stabilizes 3′ endo ribose conformation.
≥15% 2′O-MOE at the ends of the siRNA sense chain increases nuclease resistance in vitro.
|
|
Locked nucleic acid
(LNA)
|
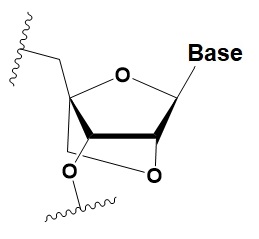
|
+2–8◦C
|
≥40% LNA in the sense chain inhibit RNAi by 5–20%.
>20% LNA in the antisense chain, or the first LNA nucleotide at the 5′ end completely inhibit RNA.
LNA can change thermal asymmetry of the duplex, increasing the efficiency of siRNA.
|
Reduces the conformational flexibility of nucleotides by fixing the C3′ endo conformation of the ribose.
≥10–20% LNA in siRNAs increase nuclease resistance in vitro and in vivo.
|
|
Bridged nucleic acid
(BNA)
|
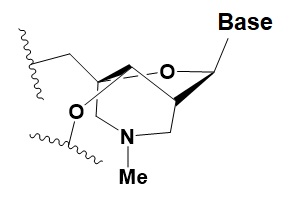
|
|
≥40% BNA in the sense chain inhibit RNAi by 8–25%.
>20% BNA in the antisense chain, or the first BNA nucleotide at the 5′ end completely inhibit RNA.
BNA changes thermal asymmetry of the duplex, increasing the efficiency of siRNA.
|
Reduces the conformational flexibility of nucleotides by fixing the C3′ endo conformation of the ribose.
≥10–20% BNA in siRNAs increase nuclease resistance in vitro and in vivo.
|
|
Unlocked nucleic acid (UNA)
|
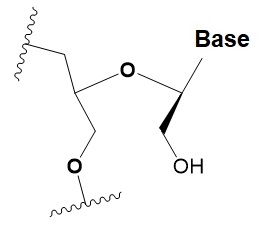
|
−5–8◦C
|
>15% UNA inhibit RNAi.
UNA can change thermal asymmetry of the duplex, increasing the efficiency of siRNA.
|
Increases conformational flexibility of nucleotides and reduces the melting point of the duplex.
UNA at the 3′ends of the duplex protect siRNA from 3′ exoribonucleases in vitro and in vivo.
|
|
4 ′ -thio-ribonucleosides (4′S)
|
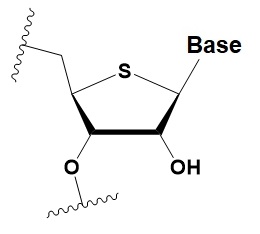
|
+1 ◦C
|
>7–15% 4′S in the antisense strand inhibit RNAi.
|
>10–15% 4′S at the ends of the strands increase the nuclease resistance in vitro.
|
|
4 ′ -C-aminomethyl-2′ -O-methyl
(4’CAm2’OMe)
|
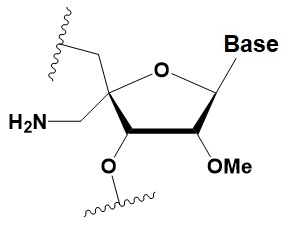
|
−1 ◦C
|
>2 analogs in the sense or >1 analog in the antisense strand inhibits RNAi.
|
≥2 modifications at the 3′ ends increase nuclease resistance in vitro.
|
|
Deoxyribonucleotide (dNMP)
|
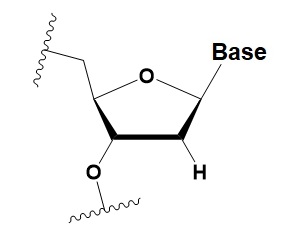
|
−0.5◦C
|
>50% dNMP inhibits RNAi.
dNMP can change thermal asymmetry of the duplex, increasing the efficiency of siRNA in vitro.
|
Protects against exoribonucleases.
|
|
Cyclohexenyl nucleic acids (CeNA)
|

|
+1.5◦C
|
5% CeNA in siRNA are tolerated by RNAi.
CeNA can change thermal asymmetry of the duplex, increasing the efficiency of siRNA in vitro.
|
Stabilizes 3′ endo ribose conformation.
≥25% CeNA analogs increase serum nuclease resistance.
|
|
Hexitol nucleic acids (HNA)
|

|
+0.85◦C
|
15% HNA in siRNA are tolerated by RNAi (Fisher et al., 2009). HNA can change thermal asymmetry of the duplex, increasing the efficiency of siRNA in vitro (Herdewijn and Juliano, 2007).
|
Slightly increases siRNA resistance to nucleases in serum (Fisher et al., 2009).
|
Adapted from: Chernikov IV, Vlassov VV, Chernolovskaya EL. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front Pharmacol. 2019 Apr 26;10:444. [PMC]
---...---