Pseudouridine (.jpg) ), one of the most abundant modified RNA molecules, is synthesized in situ by
), one of the most abundant modified RNA molecules, is synthesized in situ by .jpg) synthases. Naturally occurring RNA molecules are often modified. Over 100 different types of RNA modifications have been identified and were known for decades, however recent advances in analytical technologies have revealed that they appear to function in nearly every class of cellular RNA. It is now becoming apparent that in both coding and noncoding RNAs, dynamic modifications add a new layer of control of genetic information. Many years ago sequencing of the first biological RNA (1965) identified ten modifications including pseudouridine (
synthases. Naturally occurring RNA molecules are often modified. Over 100 different types of RNA modifications have been identified and were known for decades, however recent advances in analytical technologies have revealed that they appear to function in nearly every class of cellular RNA. It is now becoming apparent that in both coding and noncoding RNAs, dynamic modifications add a new layer of control of genetic information. Many years ago sequencing of the first biological RNA (1965) identified ten modifications including pseudouridine (.jpg) ) in the alanine transfer RNA from yeast.
) in the alanine transfer RNA from yeast.
Oligonucleotides modified with pseudouridine are useful tools to investigate how pseudouridine modified RNAs function in vivo and in vitro. Pseudouridine modified oligonucleotides can be synthesized using pseudouridine phosphoramidites and are available at Biosynthesis Inc.
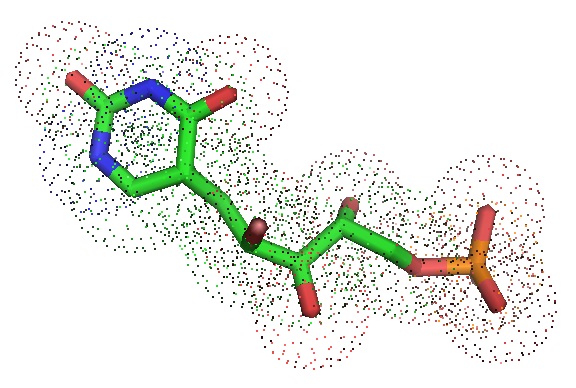
Figure 1: Structure of the ring-open pseudo-uridine mono-phosphate (ΨMP) from the crystal structure of ΨMP glycosidase/ ring-opened ribose ΨMP complex obtained by co-crystallizing ΨMP glycosidase with 4 mM MnCl2, 2 mM R5P and saturated uracil (PBD ID: 4GIL. Huang et al. 2012).
Modifications are derived from the standard nucleosides, adenosine, guanosine, cytidine and uridine. RNA modifications are present in many different types of RNA in the cell. RNA modifications are generated by enzymatic processing of the corresponding primary transcript. A large number of enzymes and pathways catalyzing post-transcriptional RNA modifications are known.

Figure 2: Conversion of uridine to pseudouridine.
Pseudouridine is synthesized by conversion of uridine. The reaction involves breaking the bond that connects the ribose to the N1 of uridine and reconnecting the ribose to position C5 of the uridine. Uridine synthases are proteins known to function without RNA cofactors. Pseudouridine is the result of isomerization or internal trans-glycosylation of uridine (U) into pseudouridine (.jpg) ), catalyzed by RNA:pseudouridine synthases. The reaction is also called a base-specific isomerization.
), catalyzed by RNA:pseudouridine synthases. The reaction is also called a base-specific isomerization.
Pseudouridine derived from isomerization of the uridine base is the most common modification in cellular RNA and abundantly found in ribosomal RNA (rRNA) and transfer RNA (tRNA). Pseudouridine is present in ~ 1 to 2 % of “salt-soluble” RNA fractions (tRNA) and < 1% in “salt-insoluble” fractions (rRNA). Recently, Pseudouridine-sequencing (PseudU-seq) established the presence of .jpg) in human and yeast mRNAs as well. A chemical labeling and pull-down method (CeU-seq) identified over
in human and yeast mRNAs as well. A chemical labeling and pull-down method (CeU-seq) identified over .jpg) 2,000 sites in human mRNA.
2,000 sites in human mRNA.
Li et al. in 2015 showed with the help of quantitative mass spectrometry that Ψ is much more prevalent (Ψ/U ratio ∼0.2-0.6%) in mammalian mRNA than previously believed. The research group developed N3-N-cyclohexyl-N’-b-(4-methyl-morpholinium) ethylcarbodiimide (CMC)-enriched pseudouridine sequencing (CeU-Seq), a selective chemical labeling and pulldown method, to identify 2,084 Ψ sites within 1,929 human transcripts, of which four (in ribosomal RNA and EEF1A1 mRNA) were biochemically verified.
Ψ modifications are dynamically regulated by the pseudouridine synthase (Pus) enzymes. Pus enzymes are known to catalyze the isomerization of Ψ in response to stress conditions such as heat shock. Ψ also affects the secondary structure of RNA. In addition, Ψ functions in altering stop codon read through which may also be biologically relevant.
Reference
Grosjean, H.; Modification and editing of RNA: historical overview and important facts to remember. In Topics in Current Genetics pp 1-22. In: Grosjean H. (eds) Fine-Tuning of RNA Functions by Modification and Editing. Topics in Current Genetics, vol 12. Springer, Berlin, Heidelberg.
Huang, S., Mahanta, N., Begley, T. P., & Ealick, S. E. (2012). Pseudouridine Monophosphate Glycosidase: a New Glycosidase Mechanism,. Biochemistry, 51(45), 9245–9255. http://doi.org/10.1021/bi3006829. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3526674/
Lewin, B.; Genes VII, Oxford University Press. 2000, pp. 715.
Li, X., Zhu, P., Ma, S., Song, J., Bai, J., Sun, F., and Yi, C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015; 11: 592–597. https://www.ncbi.nlm.nih.gov/pubmed/26075521?dopt=Abstract
Roundtree, Ian A. et al.; Dynamic RNA Modifications in Gene Expression Regulation. Cell , Volume 169 , Issue 7 , 1187 – 1200.
Yi, C., & Pan, T. (2011). Cellular Dynamics of RNA Modification. Accounts of Chemical Research, 44(12), 1380–1388. http://doi.org/10.1021/ar200057m. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3179539/
Zaringhalam M, Papavasiliou FN.; Pseudouridylation meets next-generation sequencing. Methods. 2016 Sep 1;107:63-72. doi: 10.1016/j.ymeth.2016.03.001. Epub 2016 Mar 8.
Zhao, B. S., & He, C. (2015). Pseudouridine in a new era of RNA modifications. Cell Research, 25(2), 153–154. http://doi.org/10.1038/cr.2014.143.
---...---