In eukaryotes, the 5'- terminal ends of cellular mRNAs contain an m7GpppN cap, in which N can be any nucleotide. The RNA helicase eIF4A and the scaffold protein eukaryotic translation initiation factor 4G (eIF4G) and the capping protein eIF4E are part of the complex that loads the mRNAs onto the 40 S ribosomal subunit, together with eIF3. eIF4E has a crucial role in the regulation of translation. Capping, splicing, and polyadenylation link mRNA metabolism mechanically to transcription. In addition, mRNA decay adds an additional layer of regulation to mRNA metabolism - both capping of the 5’-end and polyadenylation of the 3’-end increase mRNAs' stability.
Scientists found that RNA capping and decapping are critical features in eukaryotes. As recently discovered by Chen et al. via a method using high resolution liquid chromatography on-line mass spectrometry (LC-MS), in bacteria, the redox cofactor nicotinamide adenine dinucleotide (NAD) attaches to small regulatory RNAs in a cap-like manner. The enzyme Nudix hydrolase NudC acts as a NAD-decapping enzyme in vitro and in vivo. Cap-protein interactions can be studied and characterized with the help of LC-MS or in a combination of mass spectrometry and X-ray crystallography. For detailed structural studies of protein cap complexes, X-ray crystallography is the method of choice.
DNA ligases also require NAD as a cofactor. The superfamily of NAD+-dependent polynucleotide ligases act as repair enzyme by joining 3’-OH and 5’-PO4 DNA or RNA ends. In 2017, Unciuleac et al. reported crystal structures of the Michaelis complexes of an ATP-dependent RNA ligase (bacteriophage T4 Rnl1) and an NAD+-dependent DNA ligase (Escherichia coli LigA). The solved structures illuminate the chemical and structural basis for lysine adenylylation, via distinctive two-metal (ATP) and one-metal (NAD+) mechanisms.
.jpg)
Figure 1: Structural models of E. coli LigA (K115M) in complex with NAD+ (Unciuleac et al.; PDB ID 5TT5).
In 2016, Hoefer et al. solved the crystal structure of an Escherichia coli NAD-capped RNA hydrolase NudC substrate complex. The enzyme complex studied contained the substrates nicotinamide adenine dinucleotide (NAD) and the cleavage product nicotinamide mononucleotide (NMN). The structures revealed the catalytic residues lining the binding pocket and features of molecular recognition of substrate and product. NAD is a cofactor found in many enzymes, for example, glyceraldehyde phosphate dehydrogenase and other dehydrogenases, where it is bound by a protein domain called the Rossman fold.
The mRNA decapping enzyme NudC is a single-strand-specific RNA decapping enzyme with a strong preference for a purine as the first nucleotide. Biochemical experiments showed that NudC preferred NAD-RNA over NAD(H) by several orders of magnitude. These results suggest that NAD-RNA is its primary biological substrate. The study's findings indicate that NudC can bind a diverse cellular RNA population in an unspecific, most likely electrostatic manner.
.jpg)
Figure 2: Structural models of NudC (NADH Pyrophosphatase) in complex with NAD (5IW4). Two images of the dimeric structure are shown here. NudC is single-strand specific and has a purine preference for the 5'-terminal nucleotide and strongly prefers NAD-linked RNA (NAD-RNA) over NAD and binds to a diverse set of cellular RNAs in an unspecific manner.
The redox factor NAD is attached to small regulatory RNAs in bacteria as a cap. In Escherichia coli, the NAD cap stabilizes small regulatory RNAs in vitro against endonucleolytic processing by RNase E and against 5’-end modification by RNA pyrophosphohydrolase RppH. RNase E is involved in RNA decay, and RppH converts 5’-triphosphate RNA into 5’-monophosphate RNA triggering endonucleolytic processing. E. coli Nudix hydrolase NudC removes the NAD cap by hydrolyzing the pyrophosphate bond to produce nicotinamide mononucleotide (NMN), and 5’-monophosphate RNA. NudC is also known as NAD(H) pyrophosphohydrolase. NAD is found as a coenzyme in all living cells. NAD consists of two nucleotides joined together by their phosphate groups.
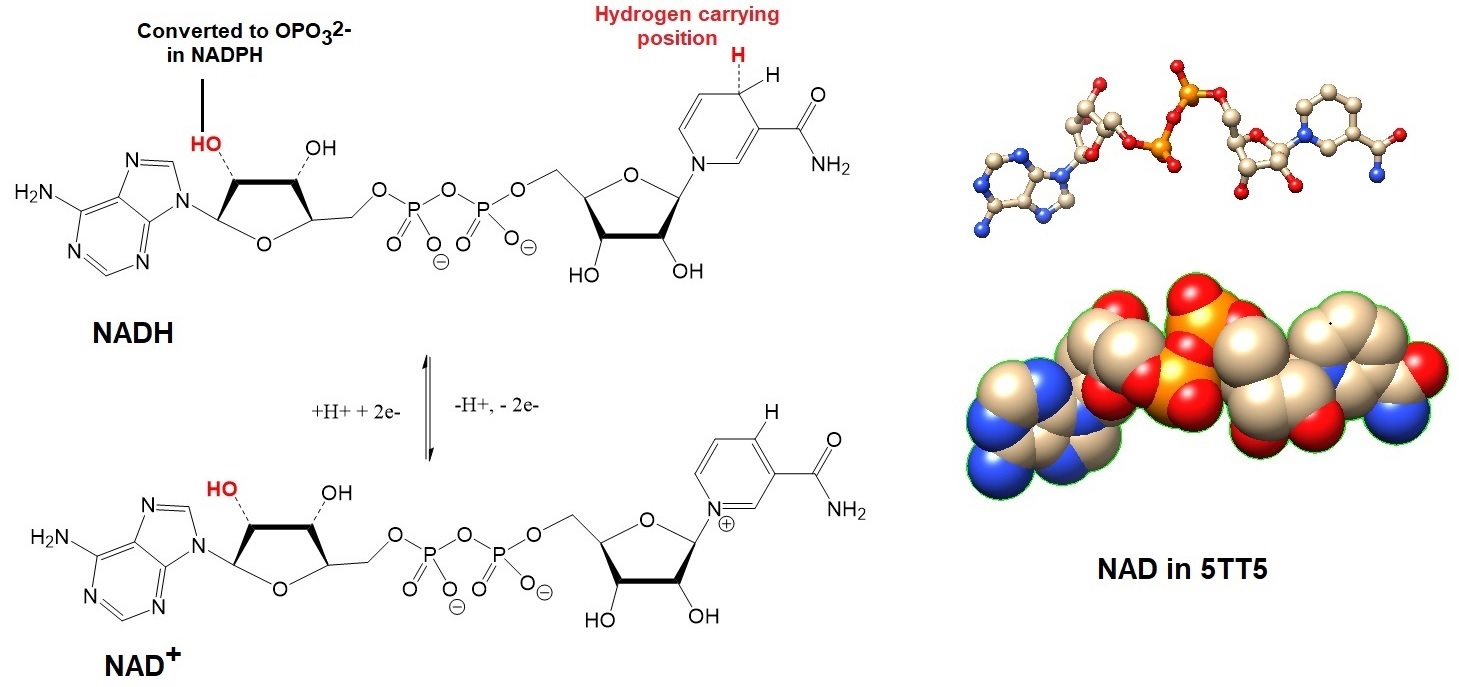
Figure 3: Structures and models for the cofactors NADH and NAD+. The hydrogen carrying cofactor NADPH is a phosphorylated form of NADH, a derivative of the vitamin nicotinamide (B3, or the water soluble form of niacin).
Not much is known about RNA processing in E. coli; however, one possibility is that NAD capping may give the bacterium additional RNA protection by using a degradation pathway that is orthogonal to the RppH-triphosphate RNA pathway.
In vitro transcription allows the addition of cap structures to RNA transcripts. A method reported by Huang F. enables the preparation of capped RNAs containing small caps including coenzyme A (CoA), flavin adenine dinucleotide (FAD), and nicotinamide adenine dinucleotide (NAD). All three coenzymes have an adenosine group. The method allows the preparation of coenzyme-linked RNA libraries useful for in vitro selection or to prepare coenzyme-coupled specific RNA sequences for other uses such as fluorescent labeling to detect specific RNA or DNA sequence, the investigation of RNA structures, and the study of RNA-RNA and RNA-protein interactions.
Adenosine derivatives such as ATP, 3’-Dephospho-coenzyme A (De-P-CoA), NAD, and FAD together with a transcription promoter sequence derived from T7 class II promoters initiate transcription. In the presence of adenosine, the preparation of adenosine-initiated RNA with free 5’-hydroxyl groups is possible. T7 RNA polymerase requires only the adenosine group to initiate transcription. Therefore, other adenosine derivatives also allow the preparation of adenosine derivative-linked RNA.
The method allows linking other biologically active molecules to the 5’-end of RNA as well including the coenzymes S-adenosylcysteine, S-adenosylhomocysteine (AdoHcy), and S-adenosylmethionine (SAM), the sugar-containing molecule adenosine 5’-diphosphoglucose (ADPG) and the signaling molecules diadenosine polyphosphates Ap(3)A and Ap(4)A.
As discovered in 1975, like eukaryotic mRNA, bacterial mRNA transcripts can also be polyadenylated.
Reference
Katherine A. Braun, Elton T. Young; Coupling mRNA Synthesis and Decay. Molecular and Cellular Biology Oct 2014, 34 (22) 4078-4087. [ https://mcb.asm.org/content/34/22/4078 ]
Brown CJ, McNae I, Fischer PM, Walkinshaw MD; Crystallographic and mass spectrometric characterisation of eIF4E with N7-alkylated cap derivatives. J Mol Biol (2007) 372 p.7-15. [PubMed]
Cap Analysis by Mass-spectrometry combined-with-X-ray-crystallography
Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009;5:879–881. [PMC] [PubMed] [Google Scholar] – A method to detect small RNA conjugates.
Höfer K, Li S, Abele F, Frindert J, Schlotthauer J, Grawenhoff J, Du J, Patel DJ, Jäschke A. Structure and function of the bacterial decapping enzyme NudC. Nat Chem Biol. 2016 Sep;12(9):730-4. [PMC]
Huang F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003 Feb 1;31(3):e8. [PMC]
Metzler, D.E.; Biochemistry. The chemical reactions of living cells. Vol 1 and 2. 2nd edition. Academic Press
Unciuleac MC, Goldgur Y, Shuman S. Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases. Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2592-2597. [PMC]
Winnacker, E-L.; From Genes to Clones. VCH.
---...---