RNAi using 2’,4’-BNANC[NH] nucleic acid analogues
RNA interference (RNAi) or gene silencing by double-stranded RNA, was discovered in 1998 and in 2006 the Nobel Prize in Physiology or Medicine was awarded to Andrew Z. Fire and Craig C. Mello for this discovery. Since then it has emerged as a vital and powerful molecular biological tool for the regulation of gene expression and is now considered to have enormous potential for the treatment of a range of diseases. Since advancements in bioengineering and nanotechnology have led to improved control of delivery and release of siRNA therapeutics, several RNAi-based therapeutics are now in Phase II and Phase III clinical trials.
RNAi is thought to be a natural defense mechanism that has evolved for the protection of organisms from RNA viruses. Cells can recognize double-stranded RNA (dsRNA) as an intruder. When this happens, the enzyme Dicer is recruited to cut the foreign RNA into smaller pieces called siRNA. These RNA pieces consist of approximately 22 nucleotides in length. One strand of the siRNA then binds to a target viral mRNA in a sequence-specific manner creating a signal for the destruction of the mRNA. The result is the interference with the further production of the viral proteins needed for a virus to replicate so that the RNA interference mechanism interferes with the expression of a particular gene that shares a sequence with the dsRNA that is homologous to that gene.
.jpg)
Mechanism of RNA interference.
RNAi technologies utilize short double-stranded RNA (dsRNA) of approximately 21 base pair length with a two nucleotide (nt) 3’-overhang for the silencing of genes. These dsRNAs are generally called small interfering RNA (siRNA). siRNA 12 to 22 nucleotides in length are the active agent in RNAi. The siRNA duplex serves as a guide for mRNA degradation. Upon siRNA incorporation into the RNA-induced silencing complex (RISC) the complex interacts with a specific mRNA and ultimately suppresses the mRNA signal. The sense strand or passenger strand of siRNA is typically cleaved at the 9th nucleotide downstream from the 5’-end of the sense strand by Argonaute 2 (Ago2) endonuclease. The activated RISC complex containing the antisense strand or guide strand binds to the target mRNA through Watson–Crick base pairing causing degradation or translational blocking of the targeted RNA.
However, the in-vivo use of RNAi or siRNA as a drug has remained difficult due to obstacles encountered such as low biostability and unacceptable toxicity possibly caused by off-target effects. Various types of chemical modifications to improve the pharmacokinetics and to overcome bio-instability problems have been investigated over the years to improve the stability and specificity of the RNAi duplexes. In some cases, the chemical modification in siRNAs has improved the serum stability of siRNAs. However, often RNAi activity was lost, but the careful placement of some specific modified residues enables enhanced siRNA biostability without loss of siRNA potency. Some of these particular modifications have reduced siRNA side effects, such as the induction of recipient immune responses and inherent off-targeting effects and have even enhanced siRNA potency. Various chemically modified siRNAs have been investigated, among them were bridged nucleic acids such as 2’,4’-methylene bridged nucleic acid 2’,4’-BNAs, also known as locked nucleic acid or LNA. Some of these modified siRNAs showed promising effects.
BNA nucleic acid analogues are useful tools for gene silencing using RNAi
Rahman et al. in 2010 investigated the effect of both 2’,4’-BNA and 2’,4’-BNA-NC modifications in gene silencing experiments using RNAi technology.
The scientists observed that 2’,4’-BNAs (LNAs) and 2’,4’-BNA-NCs are equally effective in RNAi activity if incorporated appropriately, especially in the sense strand of siRNA.
Design examples of BNA nucleic acid analogs useful for gene silencing by RNAi to inhibit the expression of firefly luciferase in CHO-luc cells. Modifications of the sense strand consecutively up to five BNA residues might be tolerable.
Native sense strand:
5’-CTTACGCUGAGUACUUCGATT-3’
Native antisense strand:
3’-TTGAAUGCGACUCAUGAAGCU-5’
Duplexes formed:
5’-CTTACGCUGAGUACUUCGATT-3’
3’-TTGAAUGCGACUCAUGAAGCU-5’
5’-GCUGAGUACUUCGAAAUGUTT-3’
3’-TTCGACUCAUGAAGCUUUACA-5’ 21mers
For the identification of the best modification, Rahman et al. investigated a range of BNAs for their ability to inhibit firefly luciferase expression in CHO-luc cells. Rahman et al. found that the introduction of 2’,4’-BNA modifications at 3’-overhangs in the sense and antisense strands of the siRNA (siBNA1 and 2) completely retained the natural RNAi property of natural RNA.
Modifications with BNAs that worked the best
5’-CTTACGCUGAGUACUUCGATT-3’ Tm = 83 °C Sense Strand
3’-TTGAAUGCGACUCAUGAAGCU-5’ Antisense Strand
5’-CTTACGCUGAGUACTUCGATT-3’ Tm = 87 °C
3’-TTGAAUGCGACUCAUGAAGCU-5’
5’-CTTACGCUGAGTACTUCGATT-3’ Tm = 86 °C
3’-TTGAAUGCGACUCAUGAAGCU-5’
5’-GCTGAGTACUTCGAAAUGTTT-3’ Tm = 86 °C
3’-TTCGACUCAUGAAGCUUUACA-5’
5’-GCTGAGTACUUCGAAATGUTT-3’ Tm = 86 °C
3’-TTCGACUCAUGAAGCUUUACA-5’
Design Rules
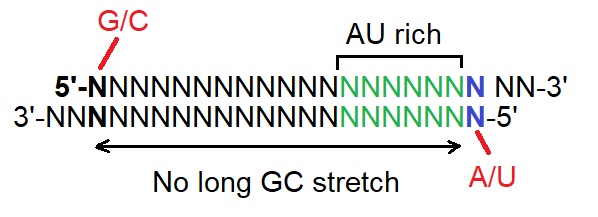
The design rules or guidlines for the design of highly effective RNAi indicate that dsRNA used for RNAi should follow the following four design parts at the same time:
(i) A/U at the 5’-end of the antisense strand,
(ii) G/C at the 5’-end of the sense strand,
(iii) AU-richness in the 5’-terminal third of the antisense strand, and
(iv) Absence of any GC stretch over 9 base pairs in length. (See figure below).
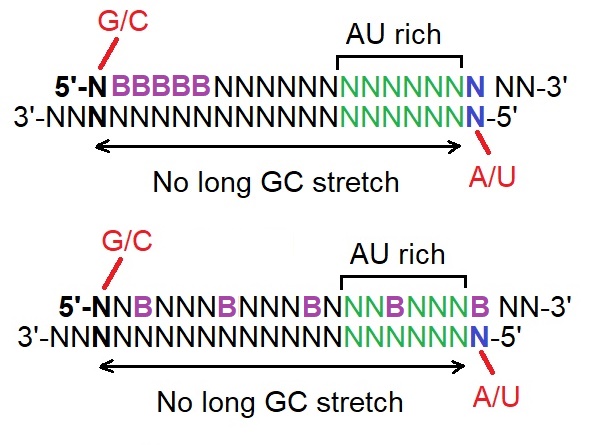
(v) Incorporation of BNAs at strategic positions.
These guidelines appear related to the molecular mechanism of RISC assembly as reported by Ui-Tei et al. in 2004. The first three guidelines together with adding BNAs to the sense strand may allow the 5’-end of the antisense strand of siRNA to be situated at or near the thermodynamically less stable siRNA duplex end.
Structures of highly effective siRNA
Please also review the following papers for more details.
Reference
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC.; Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998 Feb 19;391(6669):806-11.
S. M. Abdur Rahman, Hiroyuki Sato, Naoto Tsuda, Sunao Haitani, Keisuke Narukawa, Takeshi Imanishi, Satoshi Obika; RNA interference with 2’,4’-bridged nucleic acid analogues. Bioorganic & Medicinal Chemistry 18 (2010) 3474-3480. http://www.biosyn.com/Images/ArticleImages/pdf/RNA-interference-with-2-4-bridged-nucleic-acid-analogues.pdf, https://www.ncbi.nlm.nih.gov/pubmed/20427190
Ui-Tei,K., Naito,Y., Takahashi,F., Haraguchi,T., Ohki-Hamazaki,H., Juni,A., Ueda,R. and Saigo,K. (2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res., 32, 936–948.
---...---