What is a G-clamp?
The G-clamp, a unique molecular structure or motif, is specifically designed to interact with guanine-rich sequences in DNA or RNA oligonucleotides. It is a tricyclic analog of cytosine, the 9-(2-aminoethoxy)-phenoxazine analog known as the G-clamp. However, the 9-(3-aminopropyl)-phenoxazine analog is known as the propyl-G-clamp. The phenoxazine-derived tricyclic G-clamp strongly binds to guanine (G) through additional interactions, including π-stacking, the electrostatic attraction of a positively charged amine, and hydrogen bonding at the Hoogsteen face.
G-clamps, with their ability to selectively bind to guanine bases, find applications in various research and therapeutic settings. They are instrumental in the design of molecular probes, drugs, and other biotechnological tools. Their value in biotechnology lies in their ability to enhance the performance of oligonucleotides in a wide range of applications.
In the context of modified oligonucleotides, a "G-clamp" refers to a modified nucleic acid analog designed to increase the binding affinity and specificity for sequences containing guanine (G) bases. The "G" in G-clamp refers to guanine, one of the four nucleotides that make up DNA and RNA. The G-clamp modification enhances the stability of the hybridization between an oligonucleotide and its complementary DNA or RNA strand at sites where the guanine base is present.
In specifically designed nucleic acid analogs, a "G-clamp" modifies the affinity or specificity for guanine bases, thereby "clamping" onto guanine-containing sequences more effectively. In the antisense paradigm of drug discovery, G-clamps play a crucial role. Chemically modified oligonucleotides (ONs) are used as therapeutic agents, and G-clamps, with their improved binding to RNA sequences and stability to nucleases, contribute to their effectiveness. The identification of more potent ONs, facilitated by G-clamps, allows for the development of novel antisense-based human therapeutics that can be effective at lower doses.
G-clamps exhibit improved selective binding to guanosine (G) via (1) improved stacking interactions of the extended aromatic ring system, (2) formation of a fourth Hoogsteen-type hydrogen bond (H-bond) between a tethered amino group and O(6) of G. The G-clamp approach typically works by introducing an additional interaction, such as a hydrogen bond or a stacking interaction, with the guanine base strengthening the overall binding and making the oligonucleotide more effective in its role, whether in diagnostics, therapeutics, or research applications.
Key Applications of G-clamps are:
Antisense Oligonucleotides
In therapeutic settings, G-clamp modifications can be used to improve the efficacy of antisense oligonucleotides by increasing their binding affinity to target RNA sequences, leading to more effective gene silencing.
Probes and Primers
G-clamp modifications can enhance the sensitivity and specificity of molecular probes and primers used in techniques like PCR, qPCR, and in situ hybridization.
Gene Editing
In some gene editing technologies, G-clamp-modified oligonucleotides can help increase the precision of targeting specific DNA sequences, improving the efficiency of the editing process.
A brief history of G-Clamping
Lin & Matteucci (1998) reported a tricyclic aminoethyl-phenoxazine deoxycytidine, the so-called “G-clamp,” which tightly binds to a guanine (G) base through the formation of four hydrogen bonds in addition to a solid π-stacking effect of the tricyclic phenoxazine ring (see Figure 1).
| 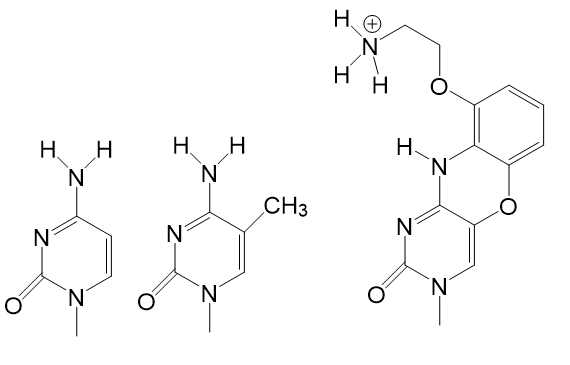 Cytosine, 5-Methyl Cytosine, Cytosine G-clamp analog. |  Model of G-clamp-guanine interactions Model of G-clamp-guanine interactions (Lin & Matteucci 1998). |
Figure 1: Chemical structures of cytosine, 5-methyl-cytosine, the cytosine G-clamp analog reported by Lin & Matteucci and the model of G-clamp-guanine interactions.
Rajeev et al. (2002) reported the synthesis of the tricyclic cytosine analogs phenoxazine, 9-(2-amino-ethoxy)-phenoxazine (G-clamp), and 9-(3-aminopropoxy)-phenoxazine (propyl-G-clamp) and their incorporation into PNA oligomers (Figure 2) using Boc-chemistry based solid-phase synthesis. PNAs with a single G-clamp modification showed a significantly enhanced affinity toward RNA and DNA targets compared to unmodified PNA with mismatch discrimination.

Figure 2: Chemical structure of a G-clamp PNA (Rajeev et al. 2002).
Mair et al. (2002) investigate the nuclease resistance of model oligomers containing a G-clamp utilizing a snake venom phosphodiesterase-based in vitro assay as the hydrolytic enzyme. The study revealed that a single incorporation at the 3'-terminus completely protected the oligonucleotides from degradation.
Wilds et el. (2003) developed an oligonucleotide analog containing a heterocycle, the guanidinium G-clamp, designed to form five H-bonds to guanosine (see Figure 3). The research group introduced the guanidinium group post-synthetically by treatment of the deprotected oligonucleotide containing a free amino group with a solution of 1H-pyrazole-1-carboxamidine. The result was that a single incorporation of this modification into an oligodeoxynucleotide sequence increased duplex stability by 13º and 16º per modification in RNA and DNA oligonucleotides.
Figure 3: H-Bonding scheme of a) G-clamp and b) guanidinium G-clamp to guanosine (Wilds et el. 2003). The introduction of G-clamp into PNA significantly improved the thermodynamic stability of the PNA/DNA duplex.
Figure 4: Structure of a DNA duplex with a single guanidinium G-clamp. (Left) PDB 1KGK: Cytosine Analog that Forms Five Hydrogen Bonds to Guanosine: the Guanidino G-Clamp. (Wilds et al. 2003). (Right) DNA duplex [d(GC*GTATMOEACGC)]2 with single guanidinium G-clamp (C*) and 2’-O-methoxyethylthymine (TMOE) modifications per strand.
Holmes et al. (2003) reported the synthesis of a 2′-O-methyl (OMe) riboside phosphoramidite derivative of the G-clamp tricyclic base and its incorporation into a series of small steric-blocking OMe oligonucleotides targeting the apical stem-loop region of human immunodeficiency virus type 1 (HIV-1) trans- activation-responsive (TAR) RNA. This study showed that an OMe G-clamp has RNA-binding enhancement abilities. The effectiveness of the oligonucleotide steric block inhibition of Tat-dependent trans-activation both in vitro and in cells is governed by factors more complex than RNA-binding strength alone. This complexity presents a fascinating challenge for further research in the field of nucleic acid chemistry and molecular biology.
Lopez-Tena et al. (2003) reported pseudo-complementary analogs of the G:C base pair leveraged on steric and electrostatic repulsion between the cationic phenoxazine analog of cytosine (G-clamp, C+) and N-7 methyl guanine (G+), both cationic. The researchers found that while complementary peptide nucleic acids (PNA) form a more stable homoduplex than the PNA:DNA heteroduplex, oligomers based on pseudo-C:G complementary PNA favor PNA:DNA hybridization. These properties enable dsDNA invasion at physiological salt concentration. The researchers obtained stable invasion complexes with low equivalents of PNAs (2–4 equiv).
This high yield of dsDNA invasion allowed the detection of RT-RPA amplicon using a lateral flow assay (LFA), which also allowed the discrimination of two strains of SARS-CoV-2 to single nucleotide resolution. Adding a G-clamp to a PNA resulted in a stronger binding affinity. However, the addition of a G-clamp resulted in a significant reduction in hybridization to N7-G but a higher affinity to unmodified G, suggesting that probes can be designed to detect this natural post-transcriptional modification by competitive binding of oligomers bearing or not a G-clamp.
Wojciechowski & Hudson (2008) developed a [bis-o-(aminoethoxy)phenyl]-pyrrolo-cytosine analog (boPhpC) designed for tight binding affinities to the guanine base and sensitive fluorescent properties. These analogs exhibit a significant increase in target-binding affinities of oligonucleotides and allow the development of sensitive fluorophore oligonucleotides.
Kuhn et al. (2010) modified a γ-PNA oligomer with a guanidinium G-clamp to enhance specific targeting of double-stranded DNA (dsDNA). The researcher group designed γ-PNAs that allow strand-invasion of dsDNA of a size length ranging from 15 to 20 base pairs. The study measured binding rates for dsDNA strand invasion by a pentadecameric γ-PNA oligomer, including targeting of matching and mismatching target sequences. The study found that synthetic γ-PNA oligomers allow selective isolation of dsDNA fragments or supercoiled plasmid DNA from DNA mixtures utilizing affinity capture protocols. The potential of γ-PNA oligomers in diagnostic assays and their versatility in other tools in molecular biology, genomics, and biotechnology opens exciting new possibilities for future research.
Also, in 2012, Ming et al. reported the synthesis of oligonucleotides containing a G-clamp or a pyrrolo-dC. This study revealed that a pyrrolo-dC derivative behaved like dC but that a G-clamp formed a more stable base pair with 2′-deoxyguanosine in DNA with parallel chain orientation than with 2′-deoxyguanosine in aps DNA.
Yamada et al. (2014) synthesized oligonucleotides with 4-N-(1H-pyrrol-2-ylcarbonyl)-deoxycytidine (dCPyc) and related derivatives. Synthetic oligodeoxynucleotides containing dCPyc hybridized with a higher affinity to DNA and RNA than the unmodified oligodeoxynucleotides. Molecular dynamic simulation studies revealed that the CPyc residue can form four hydrogen bonds with the opposite G nucleobase by keeping a more planar structure than the CInc residue where the Pyc group was replaced with a 1H-indol-2-ylcarbonyl group.
Wilds et al. (2020) also reported a "guanidino G-clamp" that can form a base pair with G using two additional hydrogen bonds.
Knizhnik et al. (2023) compared three types of such antiviral candidates: (i) locked nucleic acids (LNA), (ii) LNA–DNA gapmers, and (ii) G-clamp-containing phosphorothioates (CPSs) complementary to pseudoknot (PK) stems for their ability to unwind pseudoknot structures. The researchers found that modified antisense oligonucleotides (ONs) CPS-3 with two G-clamp insertions and total phosphorothioate backbone modifications showed only a moderate PK unfolding potential and was inferior to G54-LNA in the luciferase assays. However, the G-clamp eventually outperformed G54-LNA in cells exposed to the live virus. This result may be because, unlike LNA, CPS ONs can activate RNase H, highlighting the advantages of dual-effect antivirals.
Das et al. (2023) synthesized chlorophosphoramidate morpholino monomers containing the tricyclic cytosine analogs phenoxazine, G-clamp, and G8AE-clamp and incorporated these into 12-mer oligonucleotides. The resulting phosphorodiamidate morpholino oligomers, containing a single G-clamp, exhibited a higher affinity for complementary RNA and DNA compared to the unmodified oligomers under neutral and acidic conditions. The researchers found that duplexes of RNA and DNA with G-clamp-modified oligomers adopt a B-type helical conformation, and their binding affinities are sequence and position-dependent.
Matsubayashi et al. (2024) reported that ASOs with a G-clamp modification exhibit a high binding affinity to complementary RNA sequences. G-clamp ASOs showed efficient gene silencing effects on complementary RNAs, including MALAT1 RNA and Mapt mRNA, in vitro. This research group's in vitro studies demonstrate the promising potential of G-clamp ASOs in gene silencing, offering a positive outlook for the future of gene therapy. Also, G-clamp modifications do not alter the binding properties of proteins in cerebrospinal fluid.
López-Tena and Winssinger (2024) recently studied the impact of charges on the hybridization kinetics and thermal stability of PNA duplexes. Adding a G-clamp to a PNA oligomer results in a cationic PNA, improving the strength and stability of a PNA:DNA duplex.
Lukina et al. (2024) recently studied the recognition of 8-oxoguanosine paired with an 8-oxoG-clamp by the Human 8-oxoguanine-DNA glycosylase, the enzyme removing oxidative DNA lesions. The study showed that the oxoG-clamp fluorophore, initially proposed for the detection of oxidized purines in the DNA, impedes the removal of 8-oxoG by 8-oxoguanine-DNA-glycosylase (OGG1). OGG1 neither hydrolyzed the N-glycosidic bond in oxoguanine nor cut the sugar-phosphate backbone of the oxoG–clamp–containing DNA duplex. The oxoG-clamp residue prevents the oxoG base from protruding into the active center pocket of OGG1. Also, it hinders cleavage of abasic sites (AP: apurinic/apyrimidinic sites, the most prevalent DNA lesion sites), even if the oxidized base has already been removed. The oxoG-clamp in the complementary strand destabilizes the duplex. An OxoG-clamp presence in the opposite strand affects the thermodynamic and structural characteristics of the oligonucleotide. It changes the features of enzyme-substrate complex formation and the processing of repair enzymes and other DNA-binding proteins.
Reference
8-Oxo-G-Clamp
AP-dC-CE phosphoramidite
AP-dC = G-clamp
Das A, Ghosh A, Kundu J, Egli M, Manoharan M, Sinha S. Synthesis and Biophysical Studies of High-Affinity Morpholino Oligomers Containing G-Clamp Analogs. J Org Chem. 2023 Nov 3;88(21):15168-15175. doi: 10.1021/acs.joc.3c01658. Epub 2023 Oct 16. PMID: 37843026. [PubMed]
Efthymiou T, Gong W, Desaulniers JP. Chemical architecture and applications of nucleic acid derivatives containing 1,2,3-triazole functionalities synthesized via click chemistry. Molecules. 2012 Oct 26;17(11):12665-703. [PMC]
Flanagan WM, Wolf JJ, Olson P, Grant D, Lin KY, Wagner RW, Matteucci MD. A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc Natl Acad Sci U S A. 1999 Mar 30;96(7):3513-8. [PMC]
Guanidino G-Clamp PDB 1KGK
Haaima G., Hansen H.F., Christensen L., Dahl O., Nielsen P.E. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 1997;25:4639–4643. doi: 10.1093/nar/25.22.4639. [PMC] [PubMed]
Holmes SC, Arzumanov AA, Gait MJ. Steric inhibition of human immunodeficiency virus type-1 Tat-dependent trans-activation in vitro and in cells by oligonucleotides containing 2'-O-methyl G-clamp ribonucleoside analogues. Nucleic Acids Res. 2003 Jun 1;31(11):2759-68. [PMC]
Knizhnik E, Chumakov S, Svetlova J, Pavlova I, Khodarovich Y, Brylev V, Severov V, Alieva R, Kozlovskaya L, Andreev D, Aralov A, Varizhuk A. Unwinding the SARS-CoV-2 Ribosomal Frameshifting Pseudoknot with LNA and G-Clamp-Modified Phosphorothioate Oligonucleotides Inhibits Viral Replication. Biomolecules. 2023 Nov 17;13(11):1660. [PMC]
Kuhn H, Sahu B, Rapireddy S, Ly DH, Frank-Kamenetskii MD. Sequence specificity at targeting double-stranded DNA with a γ-PNA oligomer modified with guanidinium G-clamp nucleobases. Artif DNA PNA XNA. 2010 Jul;1(1):45-53. [PMC]
Lin, K.Y. and Matteucci, M.D., A cytosine analogue capable of clamp-like binding to guanine in helical nucleic acids. J. Am. Chem. Soc. 1998, 120, 33, 8531–8532. [ACS]
López-Tena M, Farrera-Soler L, Barluenga S, Winssinger N. Pseudo-Complementary G:C Base Pair for Mixed Sequence dsDNA Invasion and Its Applications in Diagnostics (SARS-CoV-2 Detection). JACS Au. 2023 Feb 1;3(2):449-458. [PMC]
López-Tena M, Winssinger N. Impact of charges on the hybridization kinetics and thermal stability of PNA duplexes. Org Biomol Chem. 2024 Jul 17;22(28):5759-5767. [PMC]
Lukina MV, Zhdanova PV, Koval VV. Structural and Dynamic Features of the Recognition of 8-oxoguanosine Paired with an 8-oxoG-clamp by Human 8-oxoguanine-DNA Glycosylase. Curr Issues Mol Biol. 2024 Apr 29;46(5):4119-4132. [PMC]
Maier MA, Leeds JM, Balow G, Springer RH, Bharadwaj R, Manoharan M. Nuclease resistance of oligonucleotides containing the tricyclic cytosine analogues phenoxazine and 9-(2-aminoethoxy)-phenoxazine ("G-clamp") and origins of their nuclease resistance properties. Biochemistry. 2002 Jan 29;41(4):1323-7. [PubMed]
Matsubayashi T, Yoshioka K, Lei Mon SS, Katsuyama M, Jia C, Yamaguchi T, Hara RI, Nagata T, Nakagawa O, Obika S, Yokota T. Favorable efficacy and reduced acute neurotoxicity by antisense oligonucleotides with 2',4'-BNA/LNA with 9-(aminoethoxy)phenoxazine. Mol Ther Nucleic Acids. 2024 Mar 18;35(2):102161. [PMC]
Mikame Y, Yamayoshi A. Recent Advancements in Development and Therapeutic Applications of Genome-Targeting Triplex-Forming Oligonucleotides and Peptide Nucleic Acids. Pharmaceutics. 2023 Oct 23;15(10):2515. [PMC]
Ming, X.; Ding, P.; Leonard, P.; Budow, S.; Seela, F. Parallel-stranded DNA: Enhancing duplex stability by the ‘G-clamp ’ and a pyrrolo-dC derivative. Org. Biomol. Chem. 2012, 10, 1861–1869. [RSC]
Rajeev KG, Maier MA, Lesnik EA, Manoharan M. High-affinity peptide nucleic acid oligomers containing tricyclic cytosine analogues. Org Lett. 2002 Dec 12;4(25):4395-8. [PubMed]
Wahba AS, Esmaeili A, Damha MJ, Hudson RH. A single-label phenylpyrrolocytidine provides a molecular beacon-like response reporting HIV-1 RT RNase H activity. Nucleic Acids Res. 2010 Jan;38(3):1048-56. [PMC] FRET
Wilds CJ, Maier MA, Tereshko V, Manoharan M, Egli M. Direct observation of a cytosine analogue that forms five hydrogen bonds to guanosine: guanidino G-clamp. Angew Chem Int Ed Engl. 2002 Jan 4;41(1):115-7. [PubMed]
Wilds, C.J., Maier, M.A., Manoharan, M., and Egli, M., Structural Basis for Recognition of Guanosine by a Synthetic Tricyclic Cytosine Analogue: Guanidinium G-Clamp. Helvetica 2003, 86, 966-978. [pdf]
Yamada K, Masaki Y, Tsunoda H, Ohkubo A, Seio K, Sekine M. A new modified cytosine base capable of base pairing with guanine using four hydrogen bonds. Oorg Bimol Chem. 2014 Apr 14;12(14):2255-62. [RSC]
---...---