|
|
|
Bio-Synthesis Newsletter - June 2017
|
Synthetic CRISPR RNA to minimize off-target cleavage
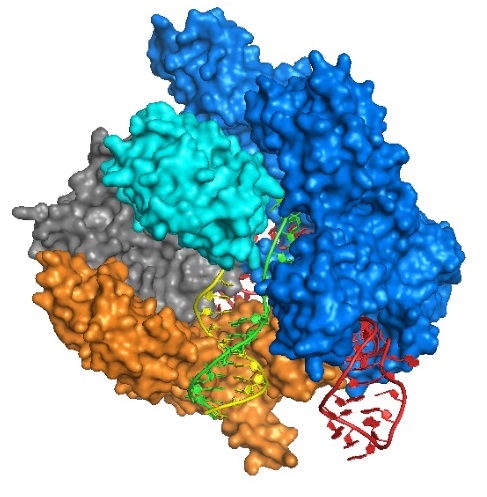 Genome editing using the CRISPR-Cas9 nuclease system is a powerful technology for manipulating genomes. This technology is now considered to be a game-changing method for the introduction of gene disruptions or corrections. In the CRISPR-Cas 9 type II system, Cas9 recognizes the complex of a 42-nucleotide CRISPR RNA (crRNA) that provides DNA specificity with the sequence adjacent to a protospacer adjacent motif (PAM) and an 80-nucleotide transactivating crRNA (tracrRNA) that binds to crRNA. Rahdar et al. recently showed that a chemically modified, 29-nucleotide synthetic crRNA could be used to replace the natural crRNA. The study showed that selective modifications of the crRNA using modified nucleotides enhances cleavage activity at the target DNA with reduced off-target cleavage. Although
various backbone modifications were used for the reported design of the synthetic crRNA bridged nucleic acids (BNAs) may also offer themselves as well for the design of CRISPR RNAs useful for therapeutic applications. Synthetic natural and modified crRNA can be commercially synthesized using standard phosphoramidite chemistry.
Genome editing using the CRISPR-Cas9 nuclease system is a powerful technology for manipulating genomes. This technology is now considered to be a game-changing method for the introduction of gene disruptions or corrections. In the CRISPR-Cas 9 type II system, Cas9 recognizes the complex of a 42-nucleotide CRISPR RNA (crRNA) that provides DNA specificity with the sequence adjacent to a protospacer adjacent motif (PAM) and an 80-nucleotide transactivating crRNA (tracrRNA) that binds to crRNA. Rahdar et al. recently showed that a chemically modified, 29-nucleotide synthetic crRNA could be used to replace the natural crRNA. The study showed that selective modifications of the crRNA using modified nucleotides enhances cleavage activity at the target DNA with reduced off-target cleavage. Although
various backbone modifications were used for the reported design of the synthetic crRNA bridged nucleic acids (BNAs) may also offer themselves as well for the design of CRISPR RNAs useful for therapeutic applications. Synthetic natural and modified crRNA can be commercially synthesized using standard phosphoramidite chemistry.
|
|
Read More
|
|
|
Circle-seq for in vitro screening of CRISPR-Cas9 nuclease targets
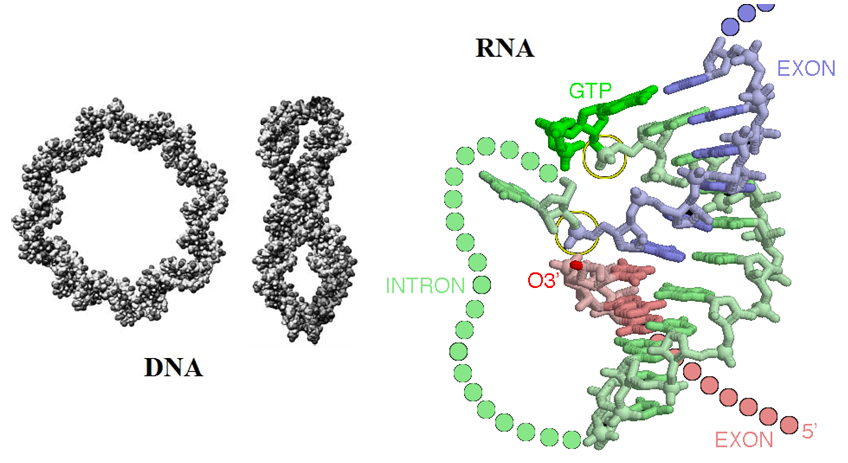 Detection of off-target effects during gene editing possibly caused by CRISPR-Cas9 nucleases is now possible using a newly developed screening strategy. CRISPR-Cas9 based gene editing methods are now considered to be game-changing methodologies. However, if CRISPR-Cas9 based gene editing methods cause off-target effects, their use for human therapeutics will be limited. Off-target effects occur due to how CRISPRs work. If more than one gene or chromosome in the genome has the same sequence string targeted by the CRISPR-Cas9 nuclease, the enzyme can cut the DNA there as well leading to a wrongly targeted part of the genome. Tsai et al. recently reported that a method called “circularization for in vitro reporting of cleavage effects by sequencing” or CIRCLE-seq is a useful tool for sensitive detection of genome-wide off-target mutations caused by CRISPR-Cas9 nucleases. Furthermore, the research group showed that CIRCLE-seq works together with next-generation sequencing technology without the need for genome sequences. Hence CIRCLE-seq allows identification of off-target mutations associated with cell-type-specific single-nucleotide polymorphisms.
Detection of off-target effects during gene editing possibly caused by CRISPR-Cas9 nucleases is now possible using a newly developed screening strategy. CRISPR-Cas9 based gene editing methods are now considered to be game-changing methodologies. However, if CRISPR-Cas9 based gene editing methods cause off-target effects, their use for human therapeutics will be limited. Off-target effects occur due to how CRISPRs work. If more than one gene or chromosome in the genome has the same sequence string targeted by the CRISPR-Cas9 nuclease, the enzyme can cut the DNA there as well leading to a wrongly targeted part of the genome. Tsai et al. recently reported that a method called “circularization for in vitro reporting of cleavage effects by sequencing” or CIRCLE-seq is a useful tool for sensitive detection of genome-wide off-target mutations caused by CRISPR-Cas9 nucleases. Furthermore, the research group showed that CIRCLE-seq works together with next-generation sequencing technology without the need for genome sequences. Hence CIRCLE-seq allows identification of off-target mutations associated with cell-type-specific single-nucleotide polymorphisms.
|
|
Read More
|
|
|
Vinylphosphonate improves siRNA-GalNac conjugates
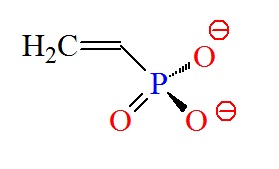 The addition of 5’-E-vinyl phosphonate (VP) to siRNA improves binding affinity to human Argonaute-2 in-vitro. The addition of VP to a trivalent N-acetylgalactosamine-siRNA conjugate enhances
gene-silencing. Efficient gene silencing using interference (RNAi ) in vivo requires recognition and binding of the 5’-phosphate of the guide strand of a siRNA by the Argonaute (Ago) protein. Unfortunately, metabolic enzymes can rapidly remove the phosphate group from the guide strand. Safe and effective siRNA delivery to the liver is enabled by conjugating a trivalent GalNac ligand to the 3’-terminus of the sense or passenger strand. To overcome instabilities, siRNAs are usually strategically modified with sugar and backbone chemical modifications such as bridged nucleic acids (BNAs) or morpholinos as well as others. Recently it was shown that vinyl phosphonate on the 5’-end of the siRNA could also be used to increase binding affinity, stability, and higher in vivo incorporation levels into hAgo2.
The addition of 5’-E-vinyl phosphonate (VP) to siRNA improves binding affinity to human Argonaute-2 in-vitro. The addition of VP to a trivalent N-acetylgalactosamine-siRNA conjugate enhances
gene-silencing. Efficient gene silencing using interference (RNAi ) in vivo requires recognition and binding of the 5’-phosphate of the guide strand of a siRNA by the Argonaute (Ago) protein. Unfortunately, metabolic enzymes can rapidly remove the phosphate group from the guide strand. Safe and effective siRNA delivery to the liver is enabled by conjugating a trivalent GalNac ligand to the 3’-terminus of the sense or passenger strand. To overcome instabilities, siRNAs are usually strategically modified with sugar and backbone chemical modifications such as bridged nucleic acids (BNAs) or morpholinos as well as others. Recently it was shown that vinyl phosphonate on the 5’-end of the siRNA could also be used to increase binding affinity, stability, and higher in vivo incorporation levels into hAgo2.
|
|
Read More
|
|
|
ATP is a hydrotrope
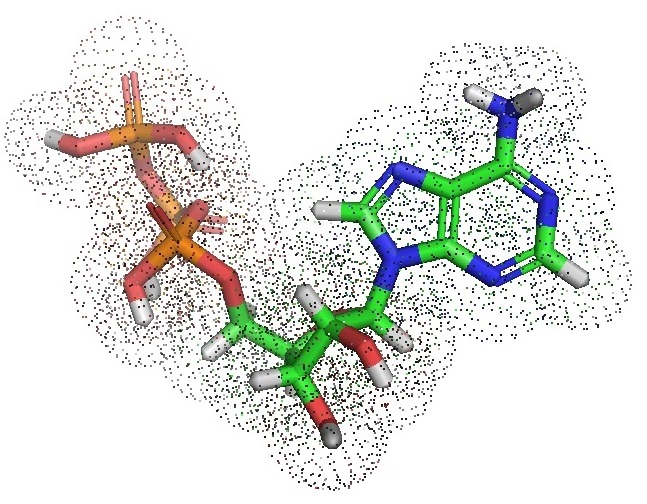 Recently Patel et al. showed that adenosine triphosphate (ATP) could prevent the formation of protein aggregates. Also, ATP can dissolve previously formed protein aggregates. Since proteins are hydrophobic molecules, ATP can be described as a biological hydrotrope. A hydrotrope is a small molecule that can solubilize hydrophobic molecules in aqueous solutions. ATP consists of adenosine containing an adenine ring, a ribose sugar and three phosphate groups referred to as the alpha (α), beta (β), and gamma (γ) phosphates. ATP is highly soluble in water, quite stable in solutions between pH 6.8 and 7.4, but rapidly hydrolyses at extreme pH ranges.
Recently Patel et al. showed that adenosine triphosphate (ATP) could prevent the formation of protein aggregates. Also, ATP can dissolve previously formed protein aggregates. Since proteins are hydrophobic molecules, ATP can be described as a biological hydrotrope. A hydrotrope is a small molecule that can solubilize hydrophobic molecules in aqueous solutions. ATP consists of adenosine containing an adenine ring, a ribose sugar and three phosphate groups referred to as the alpha (α), beta (β), and gamma (γ) phosphates. ATP is highly soluble in water, quite stable in solutions between pH 6.8 and 7.4, but rapidly hydrolyses at extreme pH ranges.
|
|
Read More
|
|
Please note that creation and use of synthetic oligos for CRISPR applications requires a license from ERS genomics, the holders of IP for CRISPR technology. We do not have a license to provide synthetic oligos used in CRISPR technology from ERSGenomics. Contact ERSGenomics.com for further information.
|
|
|
Bio-Synthesis, Inc.
800 Mario Court, Lewisville, TX 75057, USA
Toll Free: 800.227.0627 | 1.972.420.8505 (Intl.)
|
|
|
|
|
|