Bioanalytical Services
LIFE SCIENCE SERVICES
Probe & Primer Design Workflow
Design of BNANC Oligonucleotides
Artificial oligonucleotides can be used for the design of specific DNA or RNA probes !
Model of BNANC[NH] TFO with dsDNA
![Model of BNANC[NH] TFO with dsDNA Model of BNANC[NH] TFO with dsDNA](http://www.biosyn.com/images/userfiles/image/model-bna.jpg)
| CPK model of TFO |
Expanded View |
5’-d(TTTTTmCTTTmCTmCTmCT)-3’; T = BNANC[NH]
5’-d(GCTAAAAA GAAA GA GA GATCG)-3’
3’-d(CGATTTTT CTTT CT CT CTAGC)-5’ Target dsDNA
Major Techniques Needed
Electrophoresis
1D, 2D, native, reduced
Synthesis
Peptides, Oligonucleotides, Carbohydrates,Conjugates, etc.
Purifications
Peptide, Proteins, Oligonucleotides,Carbohydrates, Lipids, etc.: Various LC methods
Protein ID – DNA and RNA Sequencing
Sequencing (N-terminal, internal, C-terminal)
Mass Analysis
Protein Samples from Gels
1D, 2D, native, reduced
Analysis of Synthetic Peptide or Oligonucleotides – DNA/RNA/BNA
Peptides, Oligonucleotides, Conjugates, etc.
Analysis of purified samples
Peptide, Proteins, Oligonucleotides, Metabolites Various LC methods may be used
Protein ID
Sequencing (N-terminal, internal, C-terminal)
Center Dogma of Molecular Biology
by
FRANCIS CRICK
MRC Laboratory of Molecular Biology
Hills Road,
Cambridge CB2 2QH |
The Center Dogma of Molecular Biology deals with the detailed residue-by-residue transfer of sequential information. It states that such Information cannot be transferred from protein to either protei or nucleic acid. |
Nature Vol. 227 August 8 1970 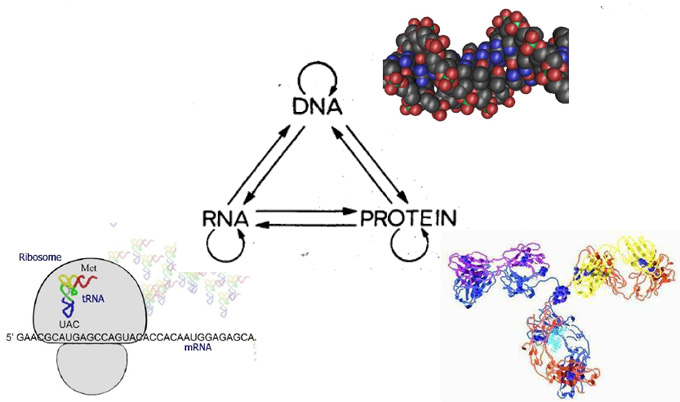
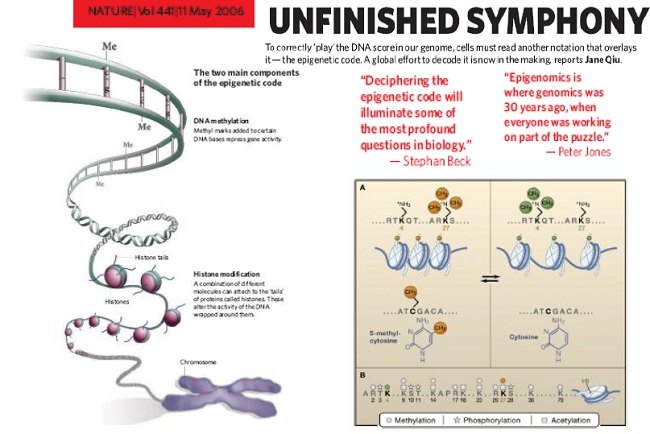
Is there a protein (histone) code?
Molecules in the cell we would like to analyze
- Proteins –catalyze reactions, form structures, control membrane permeability, cell signaling, recognize/bind other molecules, control gene function
- Nucleic acids -DNA and RNA; encode information about proteins
- Lipids - make up biomembranes
- Carbohydrates - energy sources, energy storage, constituents of nucleic acids and surface membranes
- Othersmall molecules - e.g. ATP, water, ions, etc.
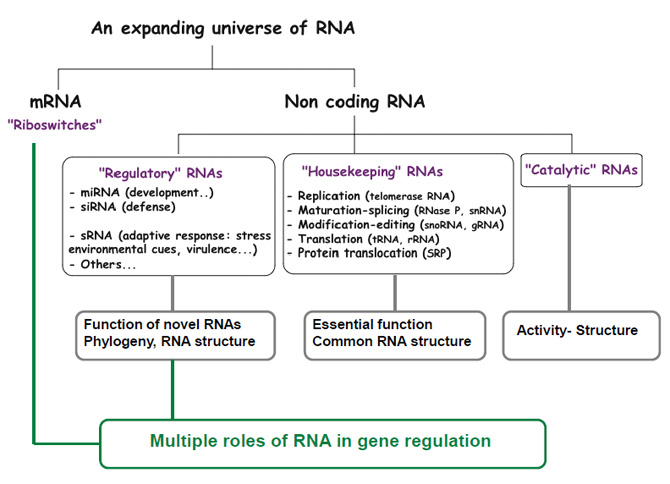
A few types of RNAs Produced in Cells
| Types of RNAs |
Functions |
| mRNAs |
messenger RNAs, code for proteins |
| rRNAs |
comprise ribosomes |
| tRNAs |
adaptors between mRNA and amino acids in protein synthesis |
| snRNAs |
splicing of pre-mRNAs |
| snoRNAs |
process and chemically modify rRNAs |
| MicroRNAs |
translation and mRNA degradation |
Other non-coding
RNAs |
telomere synthesis, X-chromo.
inactivation, protein transport |
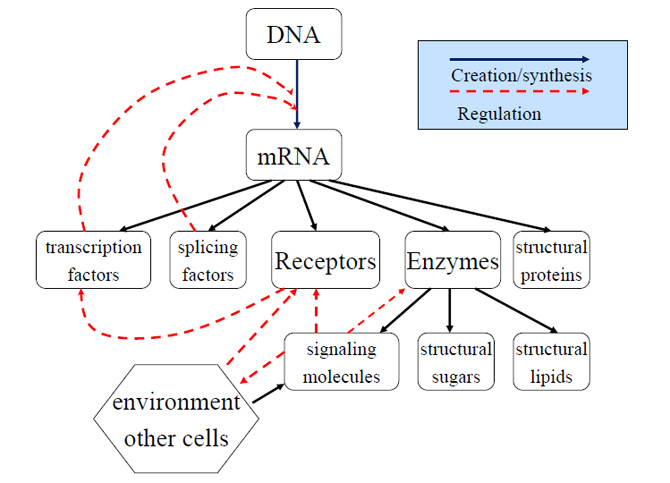
Primary Flows of Information and Substance in a Cell
Genes involved in folate metabolism
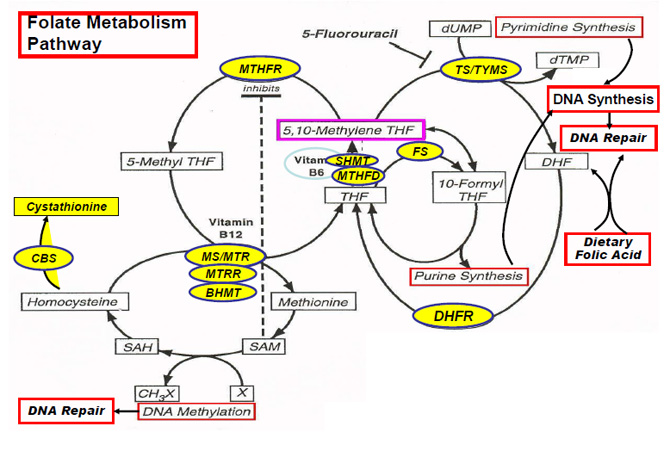
| |
MTHFR = |
methyene tetrahydrofolate transferase |
| |
TS/TYMS = |
thymidylate synthase |
| |
FS = |
10-formylTHF synthase |
| |
SHMT = |
serine hydroxymethttransferase |
| |
MTHFD |
5,10-methylenetetrahydrofolate dehydrogenase |
| |
MS/MTR = |
methionine synthase |
| |
MTRR = |
methionine synthase reductase |
| |
BHMT = |
bataine hydroxymethttransferase |
| |
DHFR = |
dihydrofolate reductase |
| |
CBS = |
systathionine Β-synthase |
| |
Modified from Ulrich CM, CEBP, 2000 |
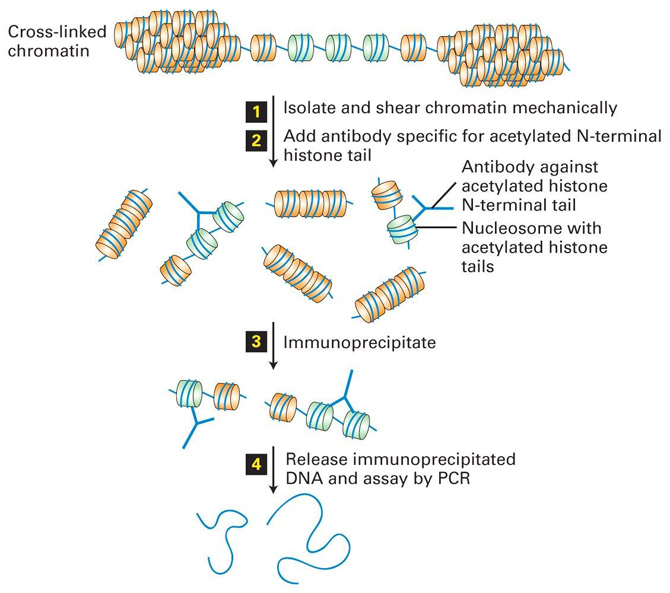
Chromatin Immunoprecipitation Reveals Histone Acetylation State
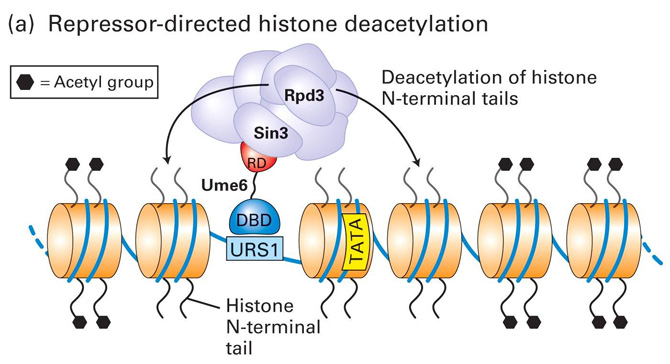
Model for Transcriptional Repression and Activation in Yeast - I
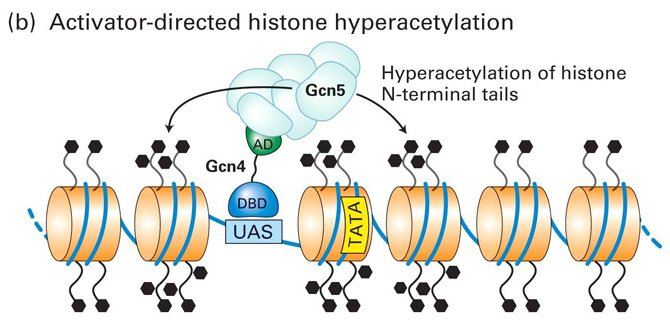
Model for Transcriptional Repression and Activation in Yeast - II
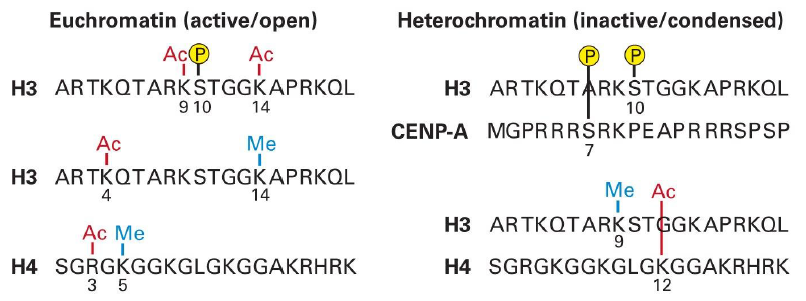
Histone Codes Control Chromatin Condensation
Proteomics – Why Now?
- We can do it now !
- Development of genome and protein sequence databases
– Bioinformatics and Data mining software
- Development of mass spectrometry instrumentation suitable to analyze biomolecules
- Protein mass, Peptide mass, Peptide sequence
- Development of analytical protein separation technology
- IEF, 2D-SDS-PAGE, HPLC, Capillary Electrophoesis, Affinity Chromatography
Sample Preparation, Pipetting, Dissolving, Digesting etc.
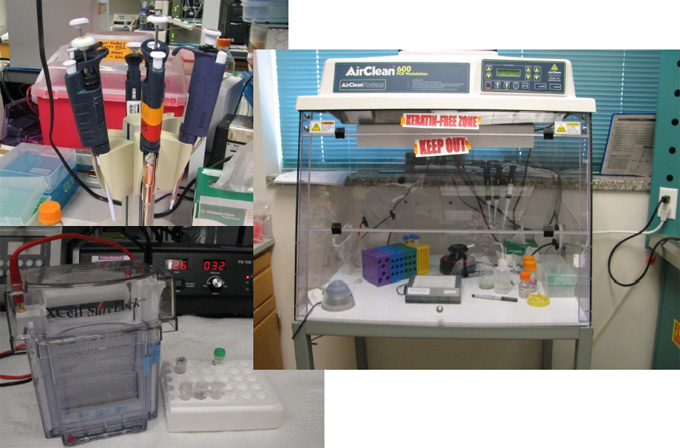
Purification of Biomolecules
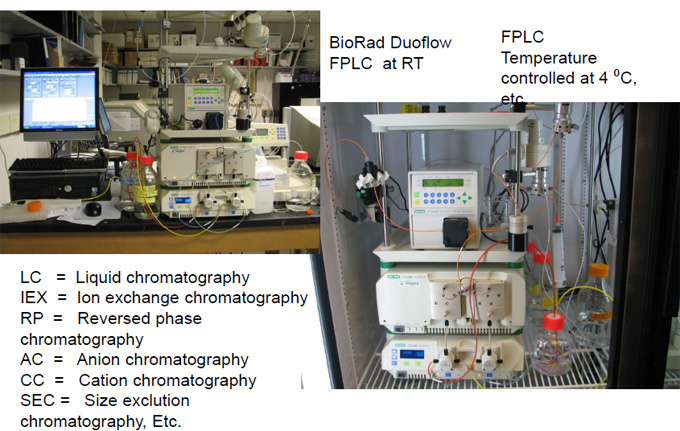
FPLC – AKTA Purifier
(Pharmacia, now GE-Amersham)

Automated HPLC

Protein Sequencing
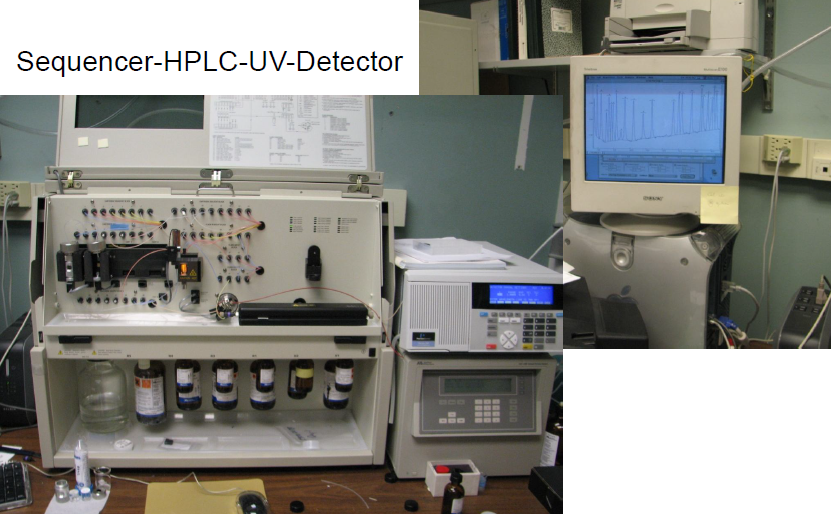
4700 MALDI-TOF-TOF MS/MS

Simple MALDI-TOF Voyager

Esquire LC-MS/MS

Protein Identification Experiment
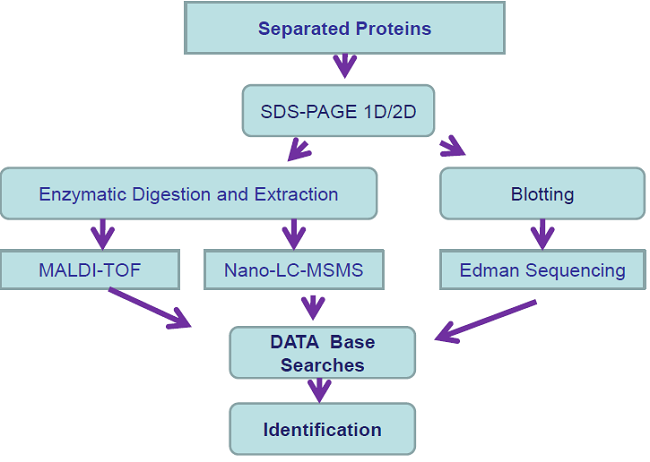
-
Primary Structure Determination
- The sequence of amino acids in a polypeptide is called its primary structure
– Several methods exist to elucidate the primary structure of peptides
Edman Degradation
- Sequential cleavage and identification of N-terminal amino acids
- Up to ~60 amino acid residues
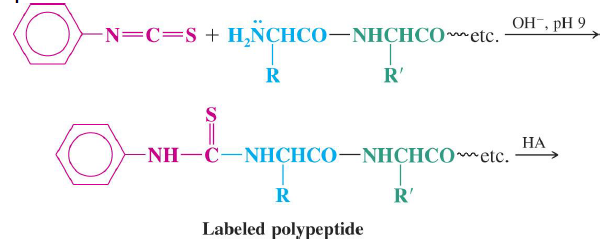
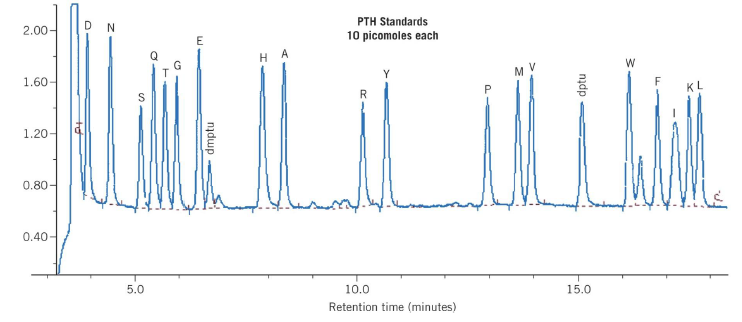
Automated amino acid sequencing
- One Edman degradation cycle beginning with a picomolar amount of polypeptide can be completed in
approximately 30 minutes
– Each cycle results in identification of the next amino acid residue in the peptide
PVDF Immobilization
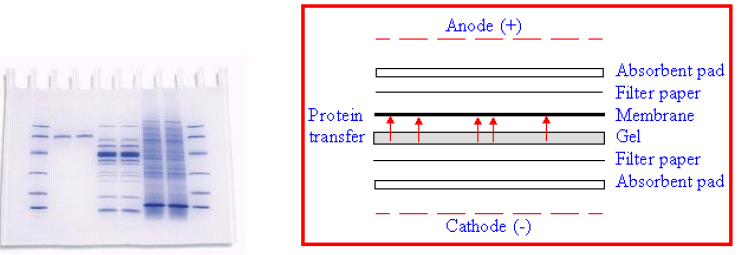
1) SDS-PAGE, 2) Electro-blotting onto PVDF
Proteins on PVDF Membrane

Protein Characterization
by
N-terminal Sequencing
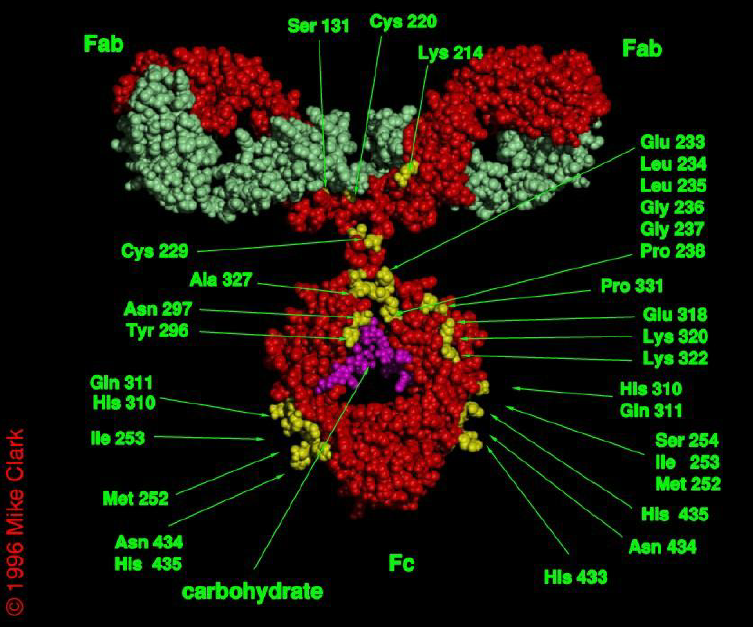
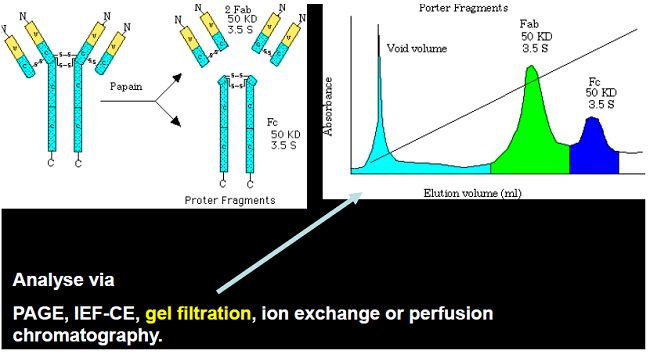
Ab Fragments Separation
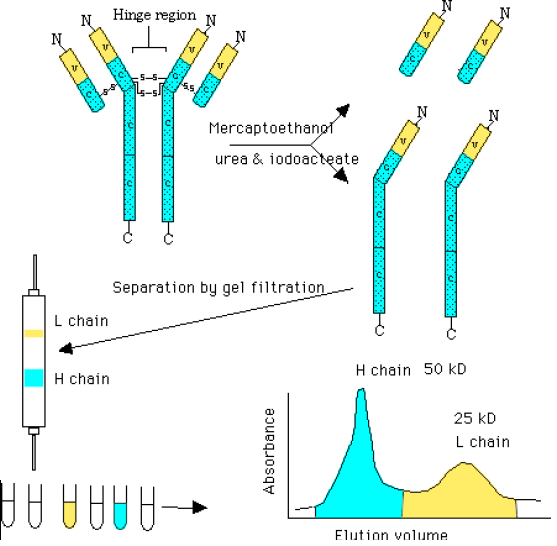
Ab Fragments after Reduction
Analysis of MABs
SDS-PAGE
Electroblotting
N-terminal Sequencing
Membrane Transfer
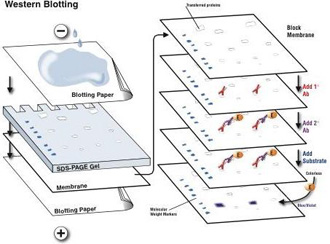 |
Transfer sandwich from cathode (-) to anode (+)
- Sponge (s)
- 3 sheets filter paper soaked in transfer buffer
- Gel
- Membrane
- 3 sheets filter paper soaked in transfer buffer
- Sponge (s)
|
Reducing PAGE & Electro-blotting of MAB’s
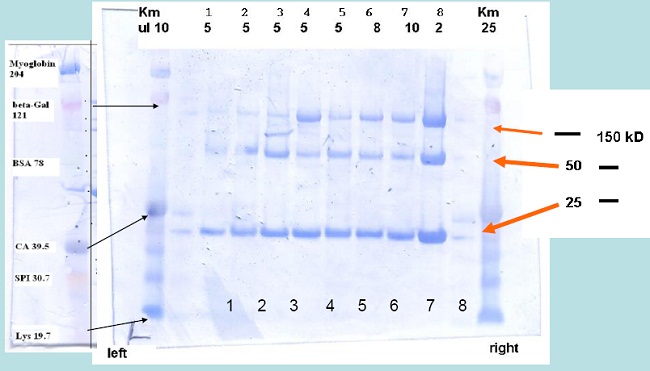 Samples were dissolved in Nupage loading buffer and reduced at 40°C for 3 h using 1 µl neat β-mercapto-ethanol
Samples were dissolved in Nupage loading buffer and reduced at 40°C for 3 h using 1 µl neat β-mercapto-ethanol
Experimental Conditions
- Reducing SDS-PAGE: NuPAGETM
4-12 % Bis-Tris Gradient Gel
- Running Buffer:
50 mM MES-Tris, 3.465 mM SDS, 1.025 mM EDTA pH 7.3, 180 to 200 V,~ 1.5 h.
- Electroblotting:
1 x NuPage transfer buffer (25 mM Bis-Tris, 25 mM Bicine, 1 mM EDTA, pH 7.2) + 20 % methanol, 1 h at 18 mA, Immobilon-P (PVDF membrane from Millipore),~ 1 h.
- Staining of PVDF membrane
15 to 30 Minutes in 50% Methanol, 10% acetic acid + comassie blue R250 (1 g/L)
- Destaining
10 Minutes to hours in 50% Methanol, 10% acetic acid.
Analysis of protein modifications
by
nano-spray LC-MSMS
Initiation of translation in procaryotes : a key step for regulation
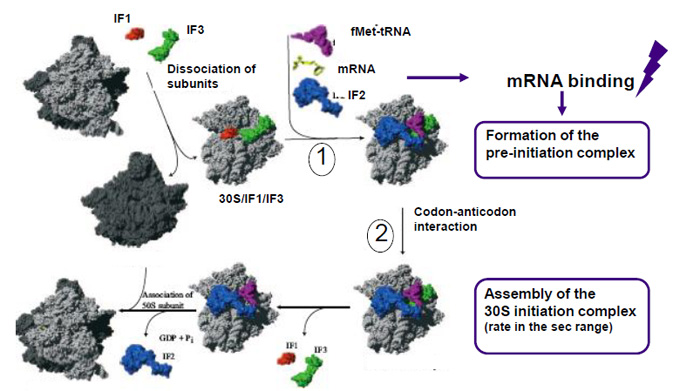
From Laursen et al. 2005 Microbiol. Mol Biol Rev. 69:101
Initiation Factor 3, IF3
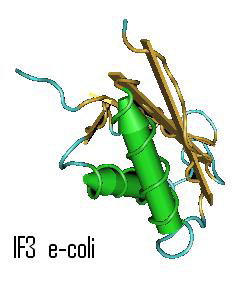 |
Garcia C, Fortier PL, Blanquet S, Lallemand JY, Dardel F
Solution structure of the ribosome-binding domain of E. coli translation initiation factor IF3. Homology with the U1A protein of the eukaryotic spliceosome J. Mol. Biol. v254, p.247-259
Initiation of translation in prokaryotes requires the formation of a complex between the messenger RNA, the 30 S ribosomal subunit and the initiator tRNA(fMet). Initiation factor IF3 binds to the 30 S ribosomal subunit and proof-reads the initiation complex, thereby ensuring the accuracy of this step. IF3 also plays a pleiotropic role in the regulation of translation, as a result of differential influences exerted on the levels of the initiation of translation of genes or groups of genes....
|
Analysis of protein from dried gels
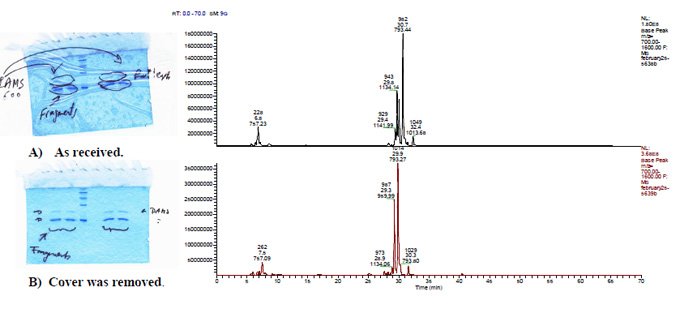
Gel bands labeled cut out and reconstituted over night in 500 ml extraction buffer (5% formic acid in 50% acetonitrile). Cellulose paper was removed. The gel bands were digested using a modified method similar to the one used by Shevchenko et al., 1996. LC-MS and LC-MS/MS analysis was performed using a Microm-LC and an LCQ deca.
translation initiation factor IF3.
1 mkggkrvqparpnrinreiraqevrltgvdgeqigivslnealekaeeagvdlveispna
61 eppvcrimdygkflyekskstkeqkeeqkviqvkeikfrpgtddgdyqvklrnlirfled
121 gdkakitlrfrgpemahqqigmevlnplrkdlcedmdlavvesfptkiegrqmimvlapk
181 kkq
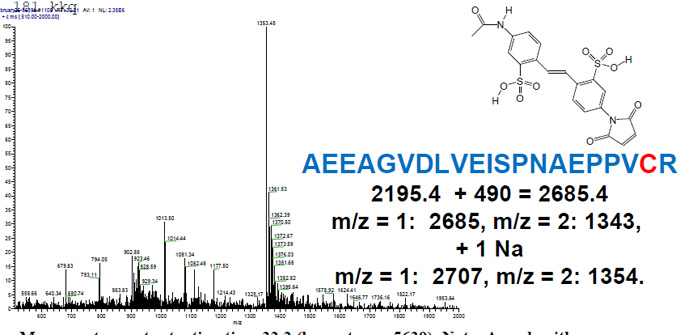
Mass spectrum at retention time 32.2 (lower trace, 5639). Note: A peak with a mass envelope from 1353 to 1381 was detected. The presence of these peak indicates that the KAEEAGVDLVEISPNAEPPVCR is labeled with the AMS fluorophor at the cysteine.
Characterization of Histones
ABRF PSRG Study
Purification
And
Analysis
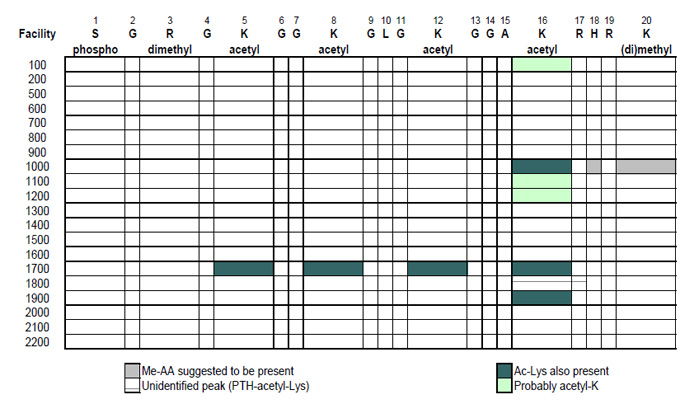
Posttranslational Modifications of Histone H4 (Bovine)
S G R G K G G K G L G K G G A K R H R K
|
FT MOD_RES 1
|
N-acetylserine
|
|
FT MOD_RES 3
|
Symmetric dimethylarginine (alternate) (By similarity)
|
|
FT MOD_RES 5
|
N6-acetyllysine (By similarity)
|
|
FT MOD_RES 8
|
N6-acetyllysine (By similarity)
|
|
FT MOD_RES 12
|
N6-acetyllysine (By similarity)
|
|
FT MOD_RES 16
|
N6-acetyllysine
|
|
FT MOD_RES 20
|
N6,N6-dimethyllysine (alternate)
|
|
FT MOD_RES 20
|
N6-methyllysine (alternate)
|
Purification
Purification of H4 (lane3) from a commercially available histone preparation (lane 2) by ion exchange and reversed-phase chromatography
PAGE
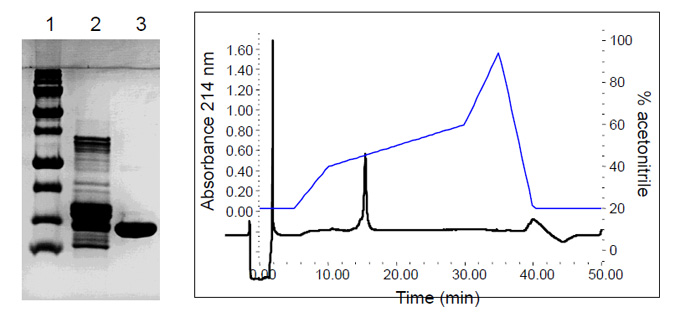
ESI MS of Bovine Histone H4
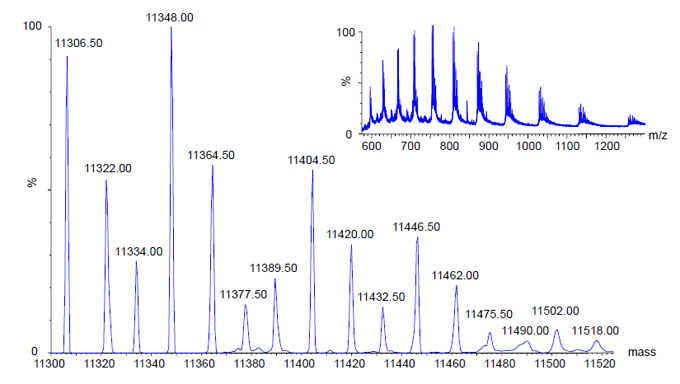
Mass Range: 575-1300 m/z ; MaxEnt: 10-13 Kd; Zoom: 11.3-11.52 Kd
Deblocking Procedures
| Procedure |
Number of Sites |
TFA/MeOH
Bergman et al. (1996) FEBS Lett. 390, 199-202 |
9 |
GAS TFA
On-Sequencer and off-line |
10 |
Liquid TFA
Wellner et al. (1990) Proc. Natl Acad. Sci. USA 87, 1947-1949 |
2 |
| BrCN/formic acid |
1 |
Species Identification
Three sites reported the identification of histone H4 for a particular species -> ?
| Arabidopsis |
1 |
MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRLAR
|
40 |
| C. elegans |
1 |
MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRLAR
|
40 |
| Yeast |
1 |
MSGRGKGGKGLGKGGAKRHRKILRDNIQGITKPAIRRLAR
|
40 |
| Bovine |
1 |
MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRLAR
|
40 |
| |
| Arabidopsis |
41 |
RGGVKRISGLIYEETRGVLKIFLENVIRDAVTYTEHARRK
|
80 |
| C. elegans |
41 |
RGGVKRISGLIYEETRGVLKVFLENVIRDAVTYCEHAKRK
|
80 |
| Yeast |
41 |
RGGVKRISGLIYEEVRAVLKSFLFSVIRDSVTYTEHAKRK
|
80 |
| Bovine |
41 |
RGGVKRISGLIYEETRGVLKVFLENVIRDAVTYTEHAKRK
|
80 |
| |
| Arabidopsis |
81 |
TVTAMDVVYALKRQGRTLYGFGG
|
103 |
| C. elegans |
81 |
TVTAMDVVYALKRQGRTLYGFGG
|
103 |
| Yeast |
81 |
TVTSLDVVYALKRQGRTLYGFGG
|
103 |
| Bovine |
81 |
TVTAMDVVYALKRQGRTLYGFGG
|
103 |
Identification of Modifications
Clostridium taeniosporum spore ribbon-1 ike appendage structure, composition and genes
|
James R. Walker,1* Annie J. Gnanam,1 Alexandra L. Bllnkova,1 Mary Jo Hermandson, 1F Mikhail A. Karymov,3 Yuri L. Lyubchenko,2 Paul R. Graves,38 Timothy A. Haysteac3 and Klaus D. Linse4
1 Molecular Genetics and Microbiology Section and institute for Cellular and Molecular Biology, University of Texas Austin, TX 78712. USA
2Department of Pharmaceutical Sciences, College of Pharmacy University of Nebraska Medical Center. Omaha, NE 68198. USA
3Department of Pharmacology, Duke University Medical Center, Durham, NC27710, USA.
4Protein Analysis Facility institute for Cellular and Molecular Biology, University of Texas, Austin, TX 78712, USA.
|
upstream of the P29a and b genes Indicate that they likely are expressed late In the mother cell, consistent with their deposition Into the layer external to the coat.
Introduction
Endospores formed by members of the Clostridium/ Bacillus subphylum typically consist of a dehydrated core containing the chromosome, a double membrane layer, a peptidoglycan cortex, and a proteinaceous coat - struc-tures which allow spore survival of extremely harsh conditions (Driks. 2003). The principal core proteins -small acid-soluble proteins - bind and saturate the DNA (Setlow. 1966; Driks. 2002a.b). The cortex is a specialized peptidoglycan. which constricts and maintains dehydra¬tion within the core (Popham etaf.: 1995; Driks: 2003).
|
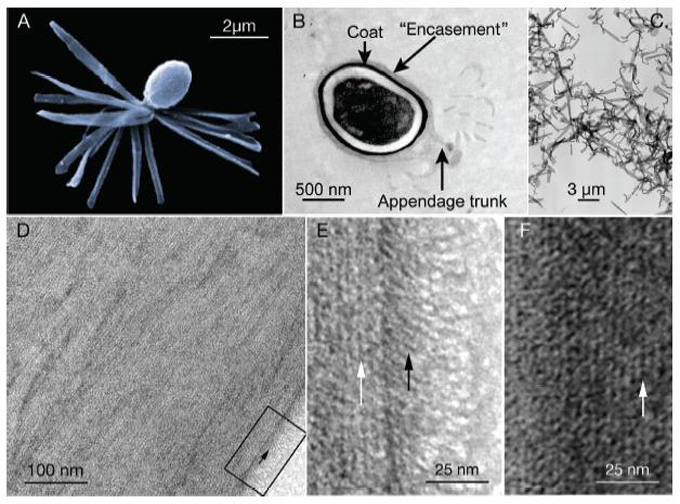 Table 2. Appendage component N-terminal and internal oligopeptide sequences.
Table 2. Appendage component N-terminal and internal oligopeptide sequences.
| A N-term ins I sequences" |
|
Present in
|
|
| MVE LKVLXSADRS YVFFGIXN |
|
P29a
|
P29b |
| MR NQYLXNRNNTG/TYND |
|
GPS5
|
|
| MKFVTNK |
|
SpoVM
|
|
| B Internal oligopeptides in P£9a and b |
|
|
|
| BNumber |
Sequence (corrected)1 |
Present in P29a
|
Present in P29b |
| P29-2 |
LDHVVFYDSLPK |
+
|
+
|
| P29-4 |
YSLSLTNIGDTK
|
+
|
-
|
| P29-S |
VFFGLSNR
|
+
|
-
|
| P29-14 |
VSYSVLLTNNSNLK |
+
|
+
|
| P29-1E |
VLTIPVIR
|
-
|
+
|
| Internal oligopeptide in GP95 |
|
|
|
|
GPS5-2
|
PPGPVGPK
|
|
|
a. X represents an unknown residue; (GVT) could be G or T
b The PES internal peptides. 4, 5 and 14 were initially sequenced as YSLSLTTLSLP, VFFGLESS and SYSVNLTNG but corrected upon re-esannination of the raw data in light of the deduced amino acid sequences of the P£9 genes. Mess spectrometry could not distinguish I and L; the final assignments were based on the deduced amino acid sequences.
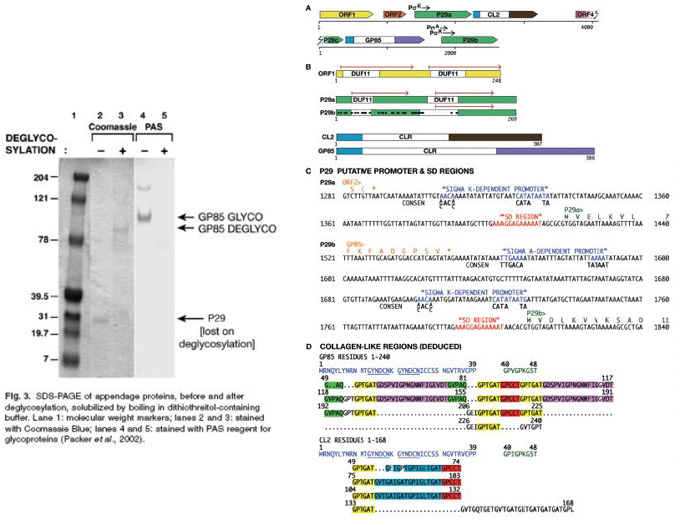
Results and Conclusions
- Integrated analysis via EM, direct protein sequencing of glycosylated and de-glycosylated coat proteins using Edman based as well as Mass Spectrometry based sequencing allowed to design primers for genomic sequencing.
- A genomic gene map of the coat protein genes could be constracted.
Common Tags for Cloning and Purification of Expressed Proteins
| His |
6 aa; HHHHHH |
Affinity |
Qiagen, Invitrogen, Roche |
|
| Step II |
8 aa; WSHPQFEK |
Affinity |
IBA |
|
| FLAG |
8 aa; DYKDDDDK |
Immunoaffinity |
Sigma |
|
| MBP |
40 kDA |
Affinity |
New England Biolabs |
Maltose binding protein |
| CBP |
4 kDa |
Affinity |
GE Healthcare |
Calmodulin binding protein epitope |
| TAB2 |
7 aa; VVSHFND |
|
Schering AG |
|
| Tab2s |
5 aa; SHFND |
|
Schering AG |
|
| c-MYC |
10 aa; EQKLISEEDL |
|
Various |
|
| HA |
9 aa; YPYDVPDYA |
Various |
|
|
| VS |
14 aa; GKPIPNPLLGLDST |
Affinity |
Invitrogen |
|
| Xpress |
8 aa; DLYDDDDK |
Affinity |
Invitrogen |
|
| BCCP |
|
|
|
Biotin Carboxyl Carrier Protein |
| Nus-tag |
495 aa; NusA protein |
Affinity |
Novagen |
|
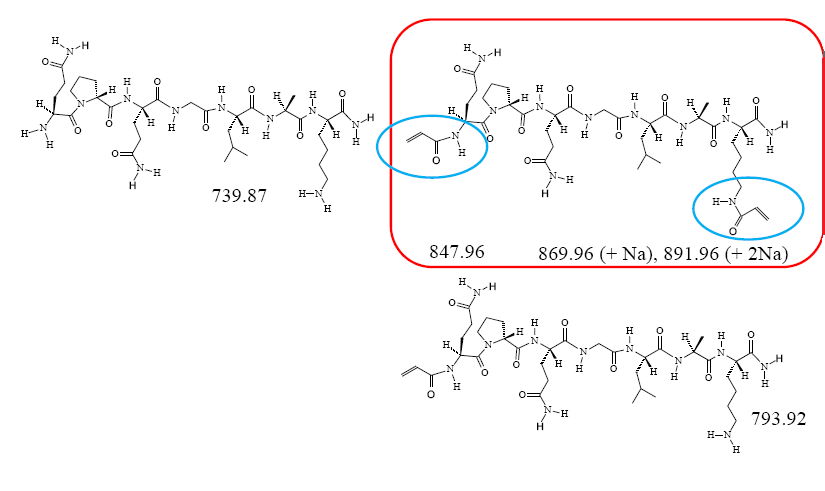
Peptide Synthesis
Fairman`& Akerfeldt, 2005
Synthesis of acryloylated Matrix Metallo Protease Peptide Precusor for Hydrogels
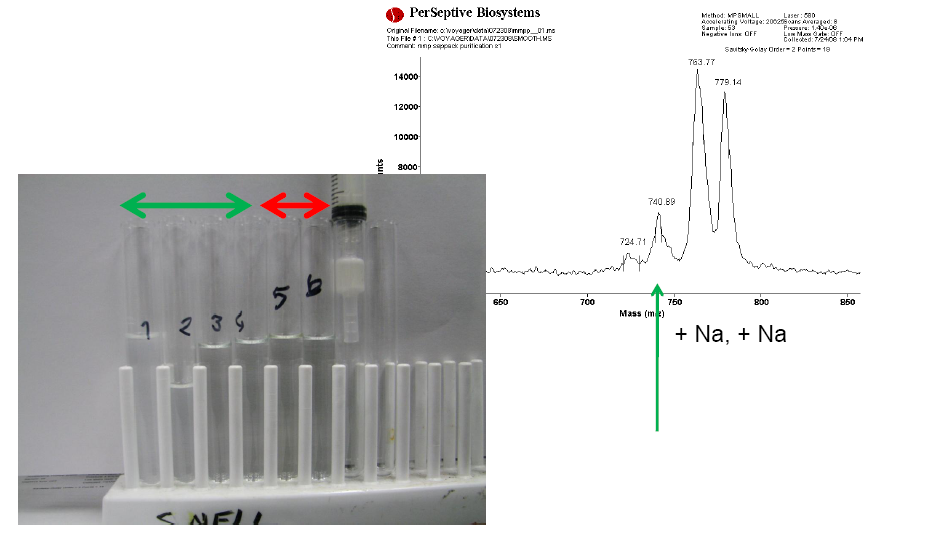
Sep-Pak Cartridge Separation
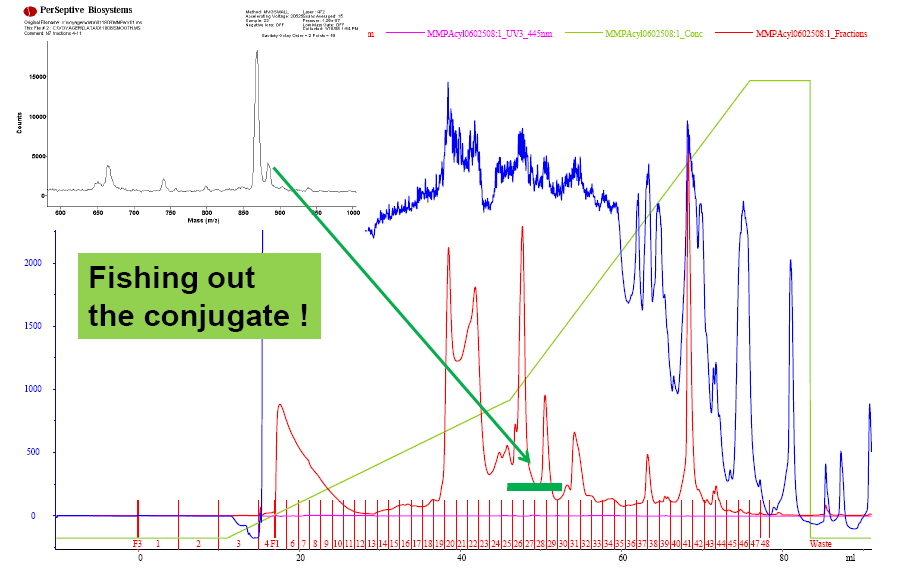
Acryloyl-MMP purification by preparative RP-HPLC
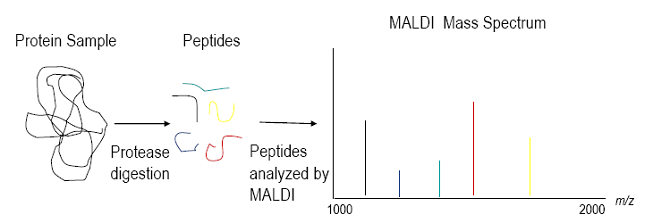
Components of Proteomics
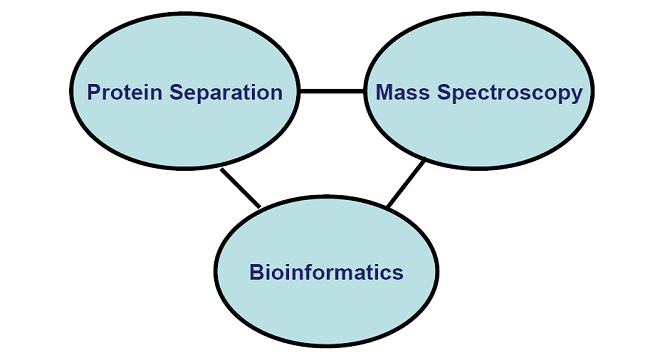
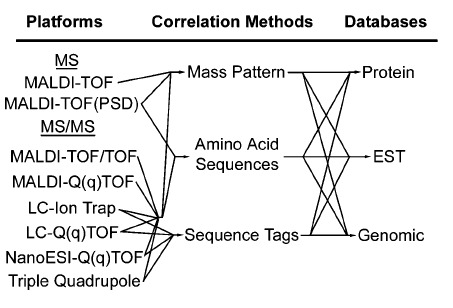
Principles of MALDI-TOF Mass Spectroscopy
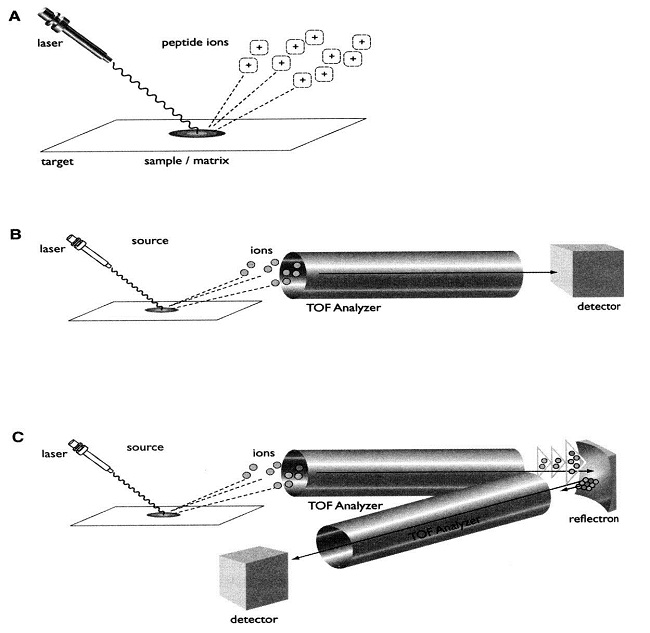
MS Technologies
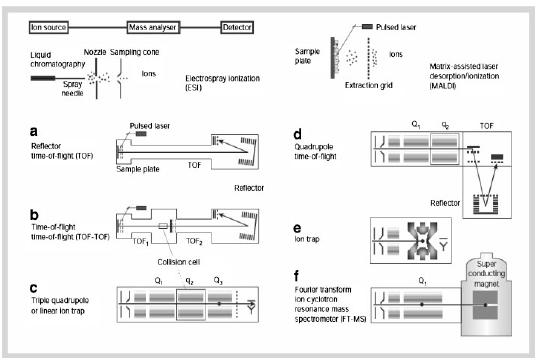
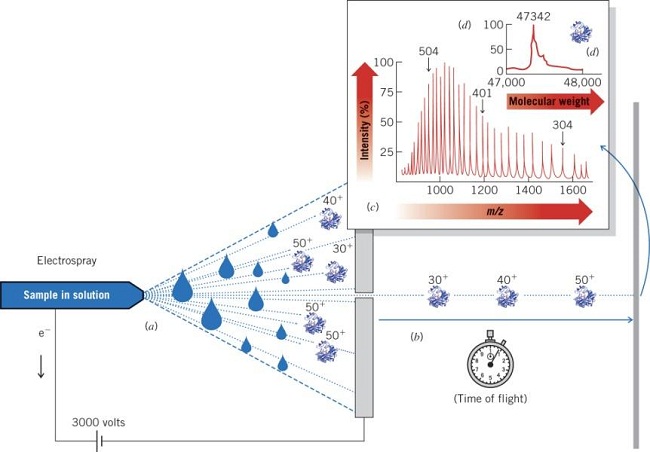
Electrospray Ionization (ESI) Mass Spectrometry (MS)
Multiply charged ions of the analyte (e.g., a protein sample) are formed by protonation in an acidic solvent One, several, or many positive charges are detected Analyte is sprayed through a high-voltage nozzle into a vacuum chamber Solvent evaporate, leaving ‘naked’ ions of the multiply charged analyte Ions are detected according to mass-to-charge (m/z) ratio Detected ions are displayed as a series according to m/z ratio Computer deconvolution of the m/z peak series leads to the molecular weight of the analyte
Peptide Mass Fingerprinting:
Search m/z Mass Tolerance (Da) # Hits Database
| 15291 |
1 |
478 |
| 1529.7 |
0.1 |
164 |
| 1529.73 |
0.01 |
25 |
| 1529.734 |
0.001 |
4 |
| 1529.7348 |
0.0001 |
2 |
|
Effect of Mass Tolerance |
| 1529.73 |
0.1 |
204 |
1529.73
1252.70 |
0.1 |
7 |
1529.73
1252.70
1833.88 |
0.1 |
1 |
|
| Effect of Multiple Peptide Masses |
| The more peptides the more stringent is the ID ! |
|
Applications of Proteomics
- Protein Mining – catalog all the proteins present in a tissue, cell, organelle, etc.
- Differential Expression Profiling – Identification of proteins in a sample as a function of a particular state: differentiation, stage of development, disease state, response to drug or stimulus
- Network Mapping – Identification of proteins in functional networks: biosynthetic pathways, signal transduction pathways, multiprotein complexes
- Mapping Protein Modifications – Characterization of posttranslational modifications: phosphorylation, glycosylation, oxidation, etc.
Why is Sequence Information Important
- A protein’s amino acid sequence is unique.
- As little as 5 amino acid sequences can ID a protein
- The sequence defines the primary structure of the protein
- the primary structure is fundamental to understanding the structure and function of the protein
- The interrelationship between an amino acid sequence and the corresponding DNA sequence
- Protein sequences access gene sequences and are key to molecular biology
|
 |
Current Methods for Proteome Research
- SDS-PAGE
- separates based on molecular weight and/or isoelectric point
- 10 fmol - > 10 pmol sensitivity
- Tracks protein expression patterns
- Protein Sequencing
- Edman degradation or internal sequence analysis
- LC-MS/MS
- Immunological Methods
Drawbacks
- SDS-Page can track the appearance, disappearance or molecular weight shifts of proteins, but can not ID the protein or measure the molecular weight with any accuracy
- Edman degradation requires a large amount of protein and does not work on N-terminal blocked proteins
- Western blotting is presumptive, requires the availability of suitable antibodies and have limited confidence in the ID related to the specificity of the antibody.
Advantageous of Mass Spectrometry
- Sensitivity in attomole range
- Rapid speed of analysis
- Ability to characterize and locate post-translational modifications
|
Papers published per year
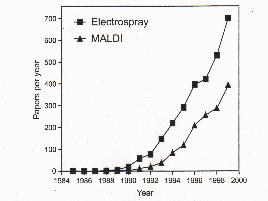 |
Enzymes for Proteome Research
|
Trypsin
|
K-X and R-X except when X = P
|
|
Endoprotease Lys-C
|
K-X except when X = P
|
|
Endoprotease Arg-C
|
R-X except when X = P
|
|
Endoprotease Asp-N
|
X-D
|
|
Endoprotease Glu-C
|
E-X except when X = P
|
|
Chymotrypsin
|
X-L, X-F, X-Y and X-W
|
|
Cyanogen Bromide
|
X-M
|
| Measured (Da) |
Theoretical (Da) |
Error |
Residues |
Sequence |
| 1381.010 |
1380.787 |
0.223 |
601-612 |
QVLLHQQALFGK |
| 1400.884 |
1400.675 |
0.209 |
337-348 |
VVWCAVGPKKQK |
| 1414.910 |
1414.752 |
0.158 |
322-333 |
NLRETAEEVKAR |
| 1505.073 |
1505.073 |
0.249 |
302-315 |
IPSKVDSALYLGSR |
| 1528.991 |
1528.991 |
0.221 |
337-349 |
VVWCAVGPEEQKK |
| 1550.985 |
1550.985 |
0.246 |
630-642 |
NLLFNDNTECLAK |
| 1725.122 |
1725.122 |
0.301 |
667-681 |
CSTSPLLEACAFLTR |
| 1827.212 |
1827.212 |
0.344 |
379-396 |
GEADALNLDGGYIYTAGK |
Micro-Sequencing by Tandem Mass Spectrometry (MS/MS)
- Ions of interest are selected in the first mass analyzer
- Collision Induced Dissociation (CID) is used to fragment the selected ions by colliding the ions with gas (typically Argon for low energy CID)
- The second mass analyzer measures the fragment ions
- The types of fragment ions observed in an MS/MS spectrum depend on many factors including primary sequence, the amount of internal energy, how the energy was introduced, charge state, etc.
- Fragmentation of peptides (amino acid chains) typically occurs along the peptide backbone. Each residue of the peptide chain successively fragments off, both in the N->C and C->N direction.
Sequence Nomenclature for Mass Ladder
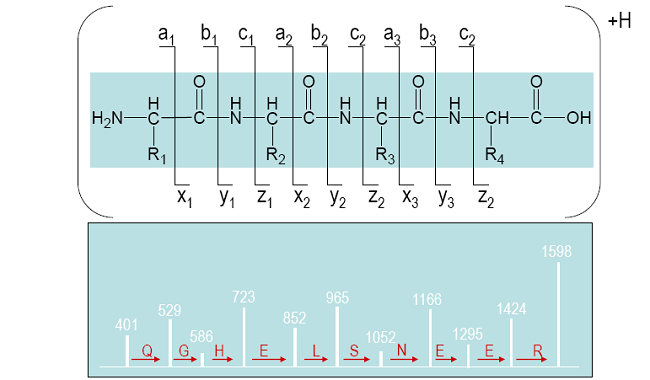
Roepstorff, P and Fohlman, J, Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom, 11(11) 601 (1984).
| |
|
|
Protein Sequence
GDVEKGKKIFVQK
CAQCHTVEKGGK
HKTGPNLHGLFG
RKTGQAPGFTYT
DANKNKGITWKE
ETLMEYLENPKKY
IPGTKMIFAGIKKK
TEREDLIAYLKKAT
NE |
|
|
Universal Proteomics Standard (UPS) Set
Catalog Number UPS1
Storage Temperature -20 °C |
|
Product Description
The Universal Proteomics Standard (UPS) Set is comprised of one vial containing 48 human source or human sequence recombinant proteins {Catalog Number U6133). and one vial (20 µg) of Proteomics Grade Trypsin (Catalog Number T6567).
There are 5 pmoles of each HPLC purified protein in the vial. Each protein has been quantitated by amino acid analysis (AAA). The proteins have been selected to limit heterogeneous post-translational modifications (PTMs).
This set can be used to standardize and/or evaluate mass spectrometry (e.g., LC-MS/MS, MALDI-TOF-MS. etc.) and electrophoretic analysis conditions prior to the analysis of complex protein samples. Moreover, the set
Sigma |
Components
Universal Proteomics Standard 1 vial
5 pmoles each of 48 human proteins, dried in a 2 ml vial Catalog Number U6133
Proteomics Grade Trypsin 20 µg
lyophilized enzyme
Catalog Number T6567
Precautions and Disclaimer
This product is for R&D use only, not for drug, household, or other uses. Please consult the Material Safety Data Sheet for information regarding hazards and safe handling practices.
Preparation Instructions |
| UniProt Accession number3 |
UniProt Protein Name [Synonym] |
MW (Da) (calculated) |
Source or recombinant |
Host |
Tag |
Potential PTMs* |
| P00709 |
Abha- lactalbumin |
14,070 |
Milk |
|
|
Glycosylation |
| P08758 |
Annexin A5 |
35,782 |
Placenta |
|
|
Acetylation |
| P01008 |
Antithrombin-lll |
49,033 |
Plasma |
|
|
Glycosylation |
| P61769 |
Beta-2-microglobulin |
11,729 |
Urine |
|
|
|
| P55957 |
BH3 interacting domain death agonist [BID] |
21,978 |
Recombinant |
E. coli |
|
|
| P00915 |
Carbonic anhydrase 1 |
28,738 |
Erythrocytes |
|
|
Acetylation |
| P00918 |
Carbonic anhydrase 2 |
29,095 |
Erythrocytes |
|
|
Acetylation |
| P04040 |
Catalase |
59,583 |
Erythrocytes |
|
|
|
| P07339 |
Cathepsin D |
26,624 |
Liver |
|
|
Glycosylation |
| P08311 |
Cathepsin G |
26,751 |
Sputum |
|
|
Glycosylation |
| P01031 |
Complement C5 [Complement C5a] |
8,266 |
Recombinant |
E. coli |
|
|
| P02741 |
C-reactive protein |
23,030 |
Plasma |
|
|
|
| P06732 |
Creatine kinase M-type [CK-MM] |
43,070 |
Heart |
|
|
|
| POO167 |
Cytochrome b5 |
16,021 |
Recombinant |
E. coli |
6-His |
|
| P99999 |
Cytochrome c [Apocytochrome c] |
11,60S |
Recombinant |
E. coh |
|
|
| P01133 |
Epidermal growth factor |
6,211 |
Recombinant |
E. coli |
|
|
| P05413 |
Fatty acid-binding protein |
14,716 |
Plasma |
|
|
Acetylation Phosphorylation |
| P06396 |
Gelsolin |
82,954 |
Plasma |
|
|
Phosphorylation |
| P08263 |
Glutathione S-transferase A1[GST A1-1] |
25,482 |
Recombinant |
E. coli |
|
|
| P09211 |
Glutathione S-transferase P [GST] |
23,220 |
Placenta |
|
|
|
| P01112 |
GTPase HRas [Ras protein] |
21,292 |
Recombinant |
E. coli |
|
|
| P69905 |
Hemoglobin alpha chain |
15,127 |
Erythrocytes |
|
|
|
| P68871 |
Hemoglobin beta chain |
15,867 |
Erythrocytes |
|
|
Acetylation,
Nitrosylation,
Glycosylation |
| P12081 |
Histidyl-tRNA synthetase [Jo-1] |
53.223 |
Recombinant |
E. coli |
|
|
| P01344 |
Insulin-like growth factor II |
7,464 |
Recombinant |
E. coli |
|
|
| P10145 |
lntefleukin-8 |
8,381 |
Recombinant |
E. coli |
|
|
| P02788 |
Lactotrans'errin |
73,289 |
Miik |
|
|
Glycosylation |
| P41159 |
Leptin |
15,024 |
Recombinant |
E. coli |
|
|
| P61626 |
LysozymeC |
14,692 |
Miik |
|
|
|
| P10636 |
Microtubule-associated prctein tau [Tau protein] |
43,810 |
Recombinant |
|
|
|
| P02144 |
Myoglobin |
17,051 |
Heart |
|
|
|
| P15559 |
NAD(P)H dehydrogenase [quinone] 1 [DT Diaphcrasej |
33,984 |
Recombinant |
E. coli |
|
|
| Q15843 |
Neddylin [Nedd8] |
9,071 |
Recombinant |
E. coli |
|
|
| P62937 |
Peptidyl-proiyl cis-trans isomerase A [Cyclophilin A] |
17,947 |
Recombinant |
E. coli |
|
|
| Q06830 |
Peroxiredcxin 1 |
22,106 |
Recombinant |
E. coli |
|
|
| P01127 |
Platelet-derived growth factor B chain |
12,286 |
Recombinant |
E. coli |
|
|
| P02753 |
Retinol-binding protein |
21,065 |
Unne |
|
|
|
| P16083 |
Ribosyidihydronicotinannde dehydrogenase (quinone) [Quinone oxidoreductase 2 or NQ02] |
25,817 |
Recombinant |
E. coli |
|
|
| P02787 |
Serotransferrin [Apotransferrin] |
75,143 |
Plasma |
|
|
Giycosylation |
| P02768 |
Serum albjmin |
65,393 |
Recombinant |
Pichia pastoris |
|
|
| P63165 |
Small ubiquitin-reiated modifier 1 [SUMO-1] |
37,420 |
Recombinant |
E. coli |
GST |
|
| P00441 |
Superoxide dismutase [Cu-Zn] |
15,800 |
Erythrocytes |
|
|
Acetylation |
| P10599 |
Thioredcxin |
12,424 |
Recombinant |
E. coli |
6-His |
|
| P01375 |
Tumor necrosis factor [TNF-afpha] |
17,350 |
Recombinant |
E. coli |
|
|
| P62988 |
Ubiquitn |
9,387 |
Recombinant |
E. coli |
6-His |
|
| P63279 |
Ubiquitin-conjugating enzyme E21 [UbcH9] |
17,995 |
Recombinant |
E. coli |
|
|
| 000762 |
Ubiquitin-conjugating enz/me E2 C [UbcHIO] |
23,473 |
Recombinant |
E. coli |
6-His |
|
| P51965 |
Ubiquitin-coniuoatino enz/me E2 E1 fUbcH61 |
22,222 |
Recombinant |
E coli |
6-His |
|
* As reported in UniProt. Potential PTMs have not been verified by Sigma.
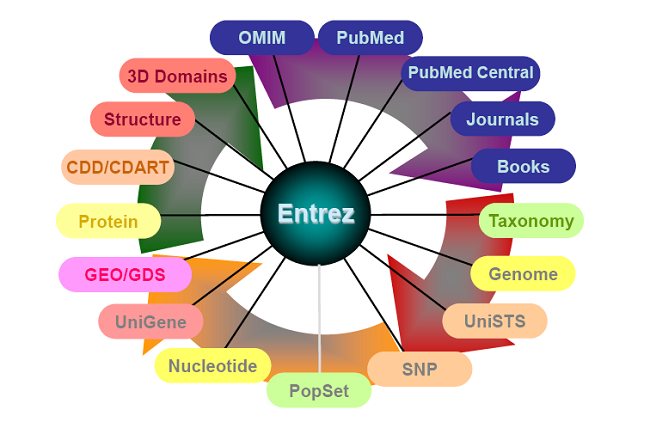
The (ever expanding) Entrez System
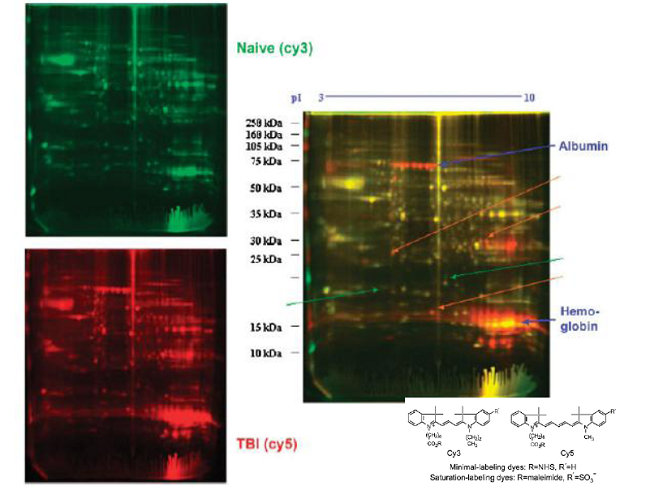
Differential gel electrophoresis 2D (DIGE)