Advantages of BNA
BNA oligonucleotides exhibit significantly higher affinity to their complementary strands when compared to previous generations of constrained nucleic acids (PNA & LNA). Their extraordinary level of sensitivity and specificity for nuclease-resistant activity makes BNA a superior tool for developing high value detection systems and therapeutic products.
Benefit of the BNA Technology
- Improved hybridization selectivity and specificity over PNA & LNA
- Ideal for the detection of short RNA and DNA targets
- Facilitate Tm normalization
- Increased thermal duplex stability of duplexes
- Increased thermal stability of triplexes
- Capable of single nucleotide discrimination
- Resistance to exo- and endonucleases resulting in high biological stability for in vivo and in vitro application
- Superior antisense inhibition and potency
- Strand invasion enable detection of "hard to access" sample
- Flexible probe designs regardless of GC content
- Easily adaptable to many DNA or RNA detection system
BNA Applications
- Antigene inhibition
- Gapmer antisense research
- In vivo, in vitro delivery
- RNAi research
- Inhibition of RNA function
- Real-time/ qPCR
- SNP detection /allele specific PCR
- In situ hybridization
- DNAzymes and Ribozymes
- Biosensor and more..
Bio-Synthesis, home of BNATM oligonucleotides
Bio-Synthesis is licensed by BNA Inc as an exclusive provider of synthetic BNA. Purchase of BNA for research use carries a "research use-only license". Clinical, therapeutic or commercial applications of BNA require a separate "commercial license".
Bridged Nucleic Acids (BNA) Technology
Natural nucleic acids have a higher degree of freedom in their chemical structure. This feature is thermodynamically unfavorable for DNA-DNA and RNA-RNA double strand formation (hybridization) and is often subject to degradation by both endo and exonucleases. Improving binding affinity (hybridizing capability) is yet unresolved for highly sensitive gene-targeting applications.
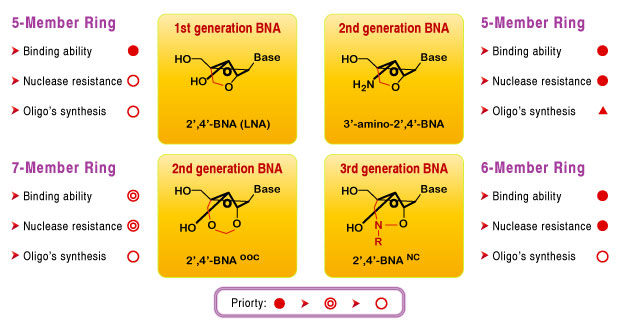
Bridged nucleic acid 2',4'-BNANC (2'-O,4'-aminoethylene bridged nucleic acid) is a compound containing a six-member bridged structure with an N-O linkage. This novel nucleic acid analogue can be synthesized and incorporated into oligonucleotides. When compared to the earlier generation of LNA, BNA was found to possess:
- Higher binding affinity against an RNA complement
- Excellent single-mismatch discriminating power
- Enhanced RNA selective binding
- Stronger and more sequence selective triplex-forming characters
- Stronger nuclease resistance to endo and exo-nucleases, even higher than the S(p)-phosphorothioateanalogue .
Based on the above results, BNA has shown great promise for applications in antisense and antigene technologies.
BNA can be easily incorporated into oligonucleotide strands. This feature allows for designing BNA hybrid oligos to satisfy the need for very high and sequence-specific hybridization with nucleic acids. Additionally, BNA possesses a strong nuclease-resistant property. Due to these outstanding properties, Bio-Synthesis Inc. now offers the third generation BNA (BNANC).
Other Type of BNA Bases
Synthesis of BNA Oligonucleotides:
BNA oligos allow greater flexibility in the design of primers and probes. They can be mixed with DNA, RNA and other nucleic acid analogs using standard phosphoramidite synthesis chemistry. BNA oligonucleotides are also easily labeled or modified with standard oligonucleotide tags such as DIG, fluorescent dyes, biotin, amino-linkers, etc.
References:
- 1.Zon, G.; Geiser, T. G., “Phosphorothioate oligonucleotides: chemistry, purification, analysis, scale-up and future directions”, Anticancer Drug Des. 1991, 6, 539-568.
- 2.Prakash, T. P.; Bhat, B., “2'-Modified oligonucleotides for antisense therapeutics”, Curr. Top. Med. Chem. 2007, 7, 641-649.
- 3.Manoharan, M., “2'-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation”, Biochim. Biophys. Acta 1999, 1489, 117-130.
- 4.Dellinger, D. J.; Sheehan, D. M.; Christensen, N. K.; Lindberg, J. G.; and. Caruthers, M. H., “Solid-phase chemical synthesis of phosphonoacetate and thiophosphonoacetate oligodeoxynucleotides”J. Am. Chem. Soc., 2003, 125, 940950.
- 5.Summerton, J., Weller, D., “Morpholino antisense oligomers: design, preparation, and properties” Antisense Nucleic Acid Drug Dev., 1997, 7:187-195.
- 6.Hyrup, B.; Nielsen, P. E., “Peptide nucleic acids (PNA): synthesis, properties and potential applications”, Bioorg. Med. Chem. 1996, 4, 5-23.
- 7.Stein, C.A; Cohen, J. S., “Oligodeoxynucleotides as inhibitors of gene expression: a review”, Cancer Res., 1988, 48, 2659-2668.
- 8.Sanghvi, Y. S., “A status update of modified oligonucleotides for chemotherapeutics applications”, Curr. Protoc. Nucleic Acid Chem., 2011, Chapter 4, Unit 4.1.1-22.
- 9.Prakash, T. P.,“An overview of sugar-modified oligonucleotides for antisense therapeutics”, Chem. Bioliers., 2011, 8, 1616-1641.
- 10.Yamamoto, T.; Nakatani, M.; Narukawa, K.; Obika, S., “Antisense drug discovery and development”, Future Med. Chem. 2011, 3, 339-365.
- 11.Veedu, R. N.; Wengel, J., “Locked nucleic acids: promising nucleic acid analogs for therapeutic applications”, Chem. Bioliers., 2010, 7, 536-542.
- 12.Rahman, S.M.; Seki, S.; Utsuki, K.; Obika, S.; Miyashita, K.; Imanishi, T., “2',4'-BNA(NC): a novel bridged nucleic acid analogue with excellent hybridizing and nuclease resistance profiles”, Nucleosides Nucleotides Nucleic Acids., 2007, 26, 1625-1628.
- 13.Rahman, S. M.; Seki, S.; Obika, S.; Yoshikawa, H.; Miyashita, K.; Imanishi, T., “Design, synthesis, and properties of 2',4'-BNA(NC): a bridged nucleic acid analogue”, J.
Enhance your applications with Bridged Nucleic Acid (BNA)
Better Tm modulation offers the most effective solution to investigate short RNA and DNA targets with extremely high sensitivity, specificity and discriminatory power1
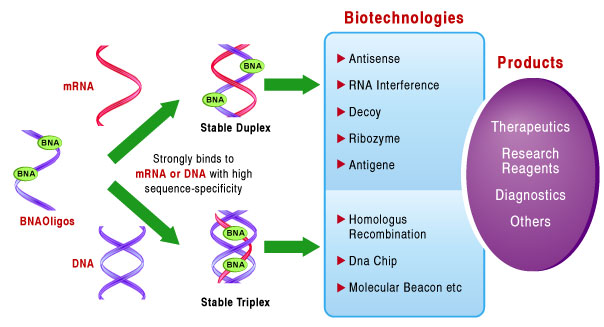
BNA monomers can be mixed within DNA, RNA, and other oligonucleotides to facilitate changes in Tm (melting temperature) without losing specificity. This feature can be used to optimize the Tm of oligonucleotides, and can shorten probe design while maintaining Tm. These are beneficial in microRNA research, PCR, microarray, and other applications where hybridization sensitivity and specificity are necessary.
Unlike its predecessor LNA, BNA possesses stronger resistance to nuclease and possesses better flexibility, both of which allow for effective duplex and triplex formation. This gives BNA oligonucleotides a high binding affinity to ssRNA and dsDNA, and makes BNA oligos an excellent tool for in-vivo applications.
Stronger biostability due to Increased thermal duplex stability and nuclease resistance, an ideal solution for in vivo antisense, siRNA application 2,3
Older generations of 2',4'-LNA do not posses sufficient resistant to nuclease, nor the flexibility required for efficient triplex formation. The increased duplex and triplex-forming ability of BNA conforms with the Watson-Crick binding and gives nBNATM oligonucleotides a high binding affinity to ssRNA and dsDNA. This strand invading property makes BNATM an excellent tool for in vivo applications. Incorporating BNA into oligonucleotides, further enhanced resistance against nuclease degradation and leads to high in vitro and in vivo stability, making BNANC a promising therapeutic agent.
Multi-functional nucleic acid analogs provide for flexibility and ease-of-use
The physiological properties of BNA such as water solubility are very similar to those of DNA and RNA. BNA can therefore be easily adapted to conventional experimental protocols, incorporated into DNA/RNA oligonucleotide synthesis, and utilized in biochemical analysis methods.
Reference:
- Miyashita, K.; Abdur Rahman, S. M.; Seki, S.; Obika, S.; Imanishi, T., "N-Methyl substituted 2',4'-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affnity RNA selective hybridization
- Yamamoto, T.; Harada-Shiba, M.; Nakatani, M.; Wada, S.; Yasuhara, H.; Narukawa, K.; Sasaki, K.; Shibata, M.-A.; Torigoe, H.; Yamaoka, T.; Imanishi, T.; Obika, S., “Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice”, Mol. Ther. Nucleic Acids, 2012, 1, e22.
- Abdur Rahman, S. M.; Sato, H.; Tsuda, N.; Haitani, S.; Narukawa, K.; Imanishi, T.; Obika, S., " RNA interference with 2',4'-bridged nucleic acid analogues", Bioorganic & Mdicinal Chemistry 18 (010) 3474-3480
BNA is a super-functional tool for nucleic acids research.
RNAi and antisense oligonucleotides
Although siRNA stimulates RNAi activity in cells, its in-vivo usage as a drug remains questionable because of low biostability and undesirable toxicity (off-target effects). To overcome these problems, nucleotide analogue usage is increasing daily.
One example is the use of Locked Nucleic Acids (LNA), the 1st generation Bridged Nucleic Acids. These have been reported to have good RNA specific binding affinity. However, LNA’s resistance to nuclease activity is significantly lower than that of phosphorothioate-containing (PS) oligonucleotides. Also, oligonucleotides containing consecutive LNA bases are very rigid, resulting in inefficient triplex formation.
Another example is Peptide nucleic acids (PNA) which also possesses promising RNA selective binding affinity. However, their mode of RNA targeting is achieved via triplex formation, which requires the use of two folds of antisense nucleic acids. In addition to that, PNA has limited aqueous solubility, poor cellular uptake, and ambiguity in binding complementary DNA/RNA in both parallel and antiparallel orientations.
Current research has shown that the nucleotide analogues 2', 4'-BNA, unlike LNA and PNA, are substantially compatible with in-vivo siRNA or antisense technology.
siBNANCoffers:
- Higher binding affinity against an RNA complement with excellent single-mismatch discriminating power,
- Much better RNA selective binding
- Stronger and more sequence selective triplex-forming characters
- Immensely higher nuclease resistance, even higher than phosphorothioate analogue
BNA oligonucleotide in DNA Research
- BNA PCR Clamp
- Real-time/quantitative PCR
- SNP detection/allele specific PCR
- In situ hybridization
- PCR clamping
- Methylation analysis
- Bead-base applications
- Chromosomal FISH
- Comparative genome hybridization
- Antigene inhibition
- Mutagenesis
- Proteomic of isolated chromatin segments (PICh)
BNA oligonucleotide in mRNA Research
- Real-time/quantitative PCR
- In situ hybridization
- Northern blotting
- Bead-base applications
- Fluorescence activated cell sorting isolation
- Inhibition of RNA function
- Microarray analysis
- RNA modification
- DNAzymes
BNA oligonucleotide in miRNA Research
- Real-time/quantitative PCR
- In situ hybridization
- Microarray analysis
- Northern blotting
- Bead-base applications
- Inhibition of RNA function
- RNA modification
- Functional analysis
BNA oligonucleotide in ncRNA Research
- Real-time/quantitative PCR
- In situ hybridization
- Fluorescence activated cell sorting
- Microarray analysis
- Northern blotting
- Inhibition of RNA function
- RNA modification
BNA oligonucleotide can also be use in
- PCR based approached
- Hybridization base approaches
- In vivo based approaches